[English] 日本語
 Yorodumi
Yorodumi- EMDB-11596: Shotgun EM of Mycobacterial protein complexes during stationary p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11596 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Shotgun EM of Mycobacterial protein complexes during stationary phase stress | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
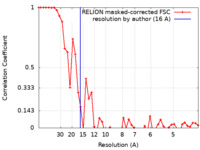
| Method | single particle reconstruction / negative staining / Resolution: 16.0 Å | |||||||||
 Authors Authors | Woodward JD / Kirykowicz AM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Res Struct Biol / Year: 2020 Journal: Curr Res Struct Biol / Year: 2020Title: Shotgun EM of mycobacterial protein complexes during stationary phase stress. Authors: Angela M Kirykowicz / Jeremy D Woodward /   Abstract: There is little structural information about the protein complexes conferring resistance in to anti-microbial oxygen and nitrogen radicals in the phagolysosome. Here, we expose the model ...There is little structural information about the protein complexes conferring resistance in to anti-microbial oxygen and nitrogen radicals in the phagolysosome. Here, we expose the model Mycobacterium, to simulated oxidative-stress conditions and apply a shotgun EM method for the structural detection of the resulting protein assemblies. We identified: glutamine synthetase I, essential for virulence; bacterioferritin A, critical for iron regulation; aspartyl aminopeptidase M18, a protease; and encapsulin, which produces a cage-like structure to enclose cargo proteins. After further investigation, we found that encapsulin carries dye-decolourising peroxidase, a protein antioxidant, as its primary cargo under the conditions tested. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11596.map.gz emd_11596.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11596-v30.xml emd-11596-v30.xml emd-11596.xml emd-11596.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11596_fsc.xml emd_11596_fsc.xml | 4.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_11596.png emd_11596.png | 49.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11596 http://ftp.pdbj.org/pub/emdb/structures/EMD-11596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11596 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11596.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11596.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacterioferritin A
| Entire | Name: Bacterioferritin A |
|---|---|
| Components |
|
-Supramolecule #1: Bacterioferritin A
| Supramolecule | Name: Bacterioferritin A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: produced by Mycobacterium smegmatis under stationary phase stress |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 440 KDa |
-Macromolecule #1: Bacterioferritin A
| Macromolecule | Name: Bacterioferritin A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: ferroxidase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MQGDPDVLKL LNEQLTSELT AINQYFLHSK MQDNWGFTEL AEHTRAESFE EMRHAETITD RILLLDGLPN YQRLFSLRVG QTLREQFEAD LAIEYEVLER LKPGIVLCRE KQDATSARLL EQILADEETH IDYLETQLQL MDKLGDALYA AQCVSRPPGS A |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl acetate | |||||||||
| Grid | Model: Homemade / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 10.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.1 kPa | |||||||||
| Details | Partially fractionated cell lysate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Number real images: 139 / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)