[English] 日本語
 Yorodumi
Yorodumi- EMDB-4407: Cryo-EM informed directed evolution of Nitrilase 4 leads to a cha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4407 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM informed directed evolution of Nitrilase 4 leads to a change in quaternary structure. | |||||||||
 Map data Map data | Synechocystis sp. PCC6803 (BAA10717.1) nitrilase truncated at residue 291 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Yamkela Q / Mulelu AE / Woodward JD | |||||||||
| Funding support |  South Africa, 1 items South Africa, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2019 Journal: Commun Biol / Year: 2019Title: Cryo-EM and directed evolution reveal how nitrilase specificity is influenced by its quaternary structure. Authors: Andani E Mulelu / Angela M Kirykowicz / Jeremy D Woodward /  Abstract: Nitrilases are helical enzymes that convert nitriles to acids and/or amides. All plants have a nitrilase 4 homolog specific for ß-cyanoalanine, while in some plants neofunctionalization has produced ...Nitrilases are helical enzymes that convert nitriles to acids and/or amides. All plants have a nitrilase 4 homolog specific for ß-cyanoalanine, while in some plants neofunctionalization has produced nitrilases with altered specificity. Plant nitrilase substrate size and specificity correlate with helical twist, but molecular details of this relationship are lacking. Here we determine, to our knowledge, the first close-to-atomic resolution (3.4 Å) cryo-EM structure of an active helical nitrilase, the nitrilase 4 from . We apply site-saturation mutagenesis directed evolution to three residues (R95, S224, and L169) and generate a mutant with an altered helical twist that accepts substrates not catalyzed by known plant nitrilases. We reveal that a loop between α2 and α3 limits the length of the binding pocket and propose that it shifts position as a function of helical twist. These insights will allow us to start designing nitrilases for chemoenzymatic synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
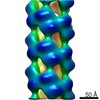
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4407.map.gz emd_4407.map.gz | 299.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4407-v30.xml emd-4407-v30.xml emd-4407.xml emd-4407.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4407.png emd_4407.png | 61.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4407 http://ftp.pdbj.org/pub/emdb/structures/EMD-4407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4407 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4407.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4407.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Synechocystis sp. PCC6803 (BAA10717.1) nitrilase truncated at residue 291 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Synechocystis sp. PCC6803 nitrilase truncated at residue 291
| Entire | Name: Synechocystis sp. PCC6803 nitrilase truncated at residue 291 |
|---|---|
| Components |
|
-Supramolecule #1: Synechocystis sp. PCC6803 nitrilase truncated at residue 291
| Supramolecule | Name: Synechocystis sp. PCC6803 nitrilase truncated at residue 291 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Synechocystis sp. PCC6803 nitrilase truncated at residue 291
| Macromolecule | Name: Synechocystis sp. PCC6803 nitrilase truncated at residue 291 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: nitrilase |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHGSHM LGKIMLNYTK NIRAAAAQIS PVLFSQQGTM EKVLDAIANA AKKGVELIVF PETFVPYYPY FSFVEPPVLM GKSHLKLYQE AVTVPGKVTQ AIAQAAKTHG MVVVLGVNER EEGSLYNTQL IFDADGALVL KRRKITPTYH ERMVWGQGDG AGLRTVDTTV ...String: HHHHHHGSHM LGKIMLNYTK NIRAAAAQIS PVLFSQQGTM EKVLDAIANA AKKGVELIVF PETFVPYYPY FSFVEPPVLM GKSHLKLYQE AVTVPGKVTQ AIAQAAKTHG MVVVLGVNER EEGSLYNTQL IFDADGALVL KRRKITPTYH ERMVWGQGDG AGLRTVDTTV GRLGALACWE HYNPLARYAL MAQHEQIHCG QFPGSMVGQI FADQMEVTMR HHALESGCFV INATGWLTAE QKLQITTDEK MHQALSGGCY TAIISPEGKH LCEPIAEGEG LAIADLDFSL IAKRKRMMDS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number grids imaged: 1 / Number real images: 50 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.1 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: PHILIPS ROTATION HOLDER |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)