+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TMV in water | |||||||||
 Map data Map data | B-factor sharpened and locally filtered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TMV / virus assembly/disassembly / Ca2+ switch / Caspar carboxylates / VIRUS | |||||||||
| Function / homology | Tobacco mosaic virus-like, coat protein / Tobacco mosaic virus-like, coat protein superfamily / Virus coat protein (TMV like) / helical viral capsid / structural molecule activity / identical protein binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Tobacco mosaic virus (strain vulgare) / Tobacco mosaic virus (strain vulgare) /  Tobacco mosaic virus (vulgare) Tobacco mosaic virus (vulgare) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||
 Authors Authors | Weis F / Beckers M | |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2019 Journal: EMBO Rep / Year: 2019Title: Elucidation of the viral disassembly switch of tobacco mosaic virus. Authors: Felix Weis / Maximilian Beckers / Iris von der Hocht / Carsten Sachse /  Abstract: Stable capsid structures of viruses protect viral RNA while they also require controlled disassembly for releasing the viral genome in the host cell. A detailed understanding of viral disassembly ...Stable capsid structures of viruses protect viral RNA while they also require controlled disassembly for releasing the viral genome in the host cell. A detailed understanding of viral disassembly processes and the involved structural switches is still lacking. This process has been extensively studied using tobacco mosaic virus (TMV), and carboxylate interactions are assumed to play a critical part in this process. Here, we present two cryo-EM structures of the helical TMV assembly at 2.0 and 1.9 Å resolution in conditions of high Ca concentration at low pH and in water. Based on our atomic models, we identify the conformational details of the disassembly switch mechanism: In high Ca /acidic pH environment, the virion is stabilized between neighboring subunits through carboxyl groups E95 and E97 in close proximity to a Ca binding site that is shared between two subunits. Upon increase in pH and lower Ca levels, mutual repulsion of the E95/E97 pair and Ca removal destabilize the network of interactions between adjacent subunits at lower radius and release the switch for viral disassembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10129.map.gz emd_10129.map.gz | 395 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10129-v30.xml emd-10129-v30.xml emd-10129.xml emd-10129.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10129.png emd_10129.png | 322.7 KB | ||
| Masks |  emd_10129_msk_1.map emd_10129_msk_1.map | 437.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10129.cif.gz emd-10129.cif.gz | 6.1 KB | ||
| Others |  emd_10129_additional.map.gz emd_10129_additional.map.gz emd_10129_half_map_1.map.gz emd_10129_half_map_1.map.gz emd_10129_half_map_2.map.gz emd_10129_half_map_2.map.gz | 16.4 MB 351 MB 351 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10129 http://ftp.pdbj.org/pub/emdb/structures/EMD-10129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10129 | HTTPS FTP |
-Related structure data
| Related structure data |  6saeMC  6sagC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10305 (Title: Cryo-EM structure of TMV in water / Data size: 6.8 EMPIAR-10305 (Title: Cryo-EM structure of TMV in water / Data size: 6.8 Data #1: Raw movies of the TMV in water sample, with corresponding start-end coordinates files and gain reference file [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10129.map.gz / Format: CCP4 / Size: 437.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10129.map.gz / Format: CCP4 / Size: 437.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened and locally filtered | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.638 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10129_msk_1.map emd_10129_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
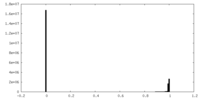
| Density Histograms |
-Additional map: Confidence map
| File | emd_10129_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Confidence map | ||||||||||||
| Projections & Slices |
| ||||||||||||
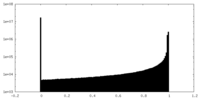
| Density Histograms |
-Half map: Halfmap 1
| File | emd_10129_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
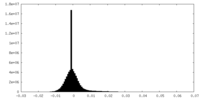
| Density Histograms |
-Half map: Halfmap 2
| File | emd_10129_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tobacco mosaic virus (strain vulgare)
| Entire | Name:  Tobacco mosaic virus (strain vulgare) Tobacco mosaic virus (strain vulgare) |
|---|---|
| Components |
|
-Supramolecule #1: Tobacco mosaic virus (strain vulgare)
| Supramolecule | Name: Tobacco mosaic virus (strain vulgare) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / NCBI-ID: 12243 / Sci species name: Tobacco mosaic virus (strain vulgare) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 Details: Residues 154-158 are flexible and were not modelled. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tobacco mosaic virus (strain vulgare) / Strain: vulgare Tobacco mosaic virus (strain vulgare) / Strain: vulgare |
| Molecular weight | Theoretical: 17.531463 KDa |
| Sequence | String: (ACE)SYSITTPSQ FVFLSSAWAD PIELINLCTN ALGNQFQTQQ ARTVVQRQFS EVWKPSPQVT VRFPDSDFKV YRYNAV LDP LVTALLGAFD TRNRIIEVEN QANPTTAETL DATRRVDDAT VAIRSAINNL IVELIRGTGS YNRSSFESSS GLVWTSG PA T UniProtKB: Capsid protein |
-Macromolecule #2: RNA (5'-R(P*GP*AP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*GP*AP*A)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Tobacco mosaic virus (vulgare) Tobacco mosaic virus (vulgare) |
| Molecular weight | Theoretical: 958.66 Da |
| Sequence | String: GAA |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 92 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 22 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: pure water |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: Pelco Easyglow, factory settings |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-20 / Number grids imaged: 1 / Number real images: 62 / Average exposure time: 5.0 sec. / Average electron dose: 30.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.35000000000000003 µm / Nominal defocus min: 0.15 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 1.406 Å Applied symmetry - Helical parameters - Δ&Phi: 22.038 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 1.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3) / Number images used: 21598 |
|---|---|
| Segment selection | Number selected: 21709 / Software - Name: SPRING |
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 3) |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)