+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6sae | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TMV in water | ||||||
 Components Components |
| ||||||
 Keywords Keywords | VIRUS / TMV / virus assembly/disassembly / Ca2+ switch / Caspar carboxylates | ||||||
| Function / homology |  Function and homology information Function and homology informationhelical viral capsid / structural molecule activity / identical protein binding Similarity search - Function | ||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | ||||||
| Method | ELECTRON MICROSCOPY / helical reconstruction / cryo EM / Resolution: 1.9 Å | ||||||
 Authors Authors | Weis, F. / Beckers, M. / Sachse, C. | ||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2019 Journal: EMBO Rep / Year: 2019Title: Elucidation of the viral disassembly switch of tobacco mosaic virus. Authors: Felix Weis / Maximilian Beckers / Iris von der Hocht / Carsten Sachse /  Abstract: Stable capsid structures of viruses protect viral RNA while they also require controlled disassembly for releasing the viral genome in the host cell. A detailed understanding of viral disassembly ...Stable capsid structures of viruses protect viral RNA while they also require controlled disassembly for releasing the viral genome in the host cell. A detailed understanding of viral disassembly processes and the involved structural switches is still lacking. This process has been extensively studied using tobacco mosaic virus (TMV), and carboxylate interactions are assumed to play a critical part in this process. Here, we present two cryo-EM structures of the helical TMV assembly at 2.0 and 1.9 Å resolution in conditions of high Ca concentration at low pH and in water. Based on our atomic models, we identify the conformational details of the disassembly switch mechanism: In high Ca /acidic pH environment, the virion is stabilized between neighboring subunits through carboxyl groups E95 and E97 in close proximity to a Ca binding site that is shared between two subunits. Upon increase in pH and lower Ca levels, mutual repulsion of the E95/E97 pair and Ca removal destabilize the network of interactions between adjacent subunits at lower radius and release the switch for viral disassembly. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6sae.cif.gz 6sae.cif.gz | 101.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6sae.ent.gz pdb6sae.ent.gz | 79.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6sae.json.gz 6sae.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sa/6sae https://data.pdbj.org/pub/pdb/validation_reports/sa/6sae ftp://data.pdbj.org/pub/pdb/validation_reports/sa/6sae ftp://data.pdbj.org/pub/pdb/validation_reports/sa/6sae | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  10129MC  6sagC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10305 (Title: Cryo-EM structure of TMV in water / Data size: 6.8 EMPIAR-10305 (Title: Cryo-EM structure of TMV in water / Data size: 6.8 Data #1: Raw movies of the TMV in water sample, with corresponding start-end coordinates files and gain reference file [micrographs - multiframe]) |
- Links
Links
- Assembly
Assembly
| Deposited unit | 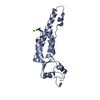
|
|---|---|
| 1 | x 40
|
| Number of models | 3 |
- Components
Components
| #1: Protein | Mass: 17531.463 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: Residues 154-158 are flexible and were not modelled. Source: (natural)  Tobacco mosaic virus (strain vulgare) / Strain: vulgare / References: UniProt: P69687 Tobacco mosaic virus (strain vulgare) / Strain: vulgare / References: UniProt: P69687 | ||||||
|---|---|---|---|---|---|---|---|
| #2: RNA chain | Mass: 958.660 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Tobacco mosaic virus (vulgare) Tobacco mosaic virus (vulgare) | ||||||
| #3: Chemical | ChemComp-MG / #4: Water | ChemComp-HOH / | Has ligand of interest | N | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: HELICAL ARRAY / 3D reconstruction method: helical reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Tobacco mosaic virus (strain vulgare) / Type: VIRUS / Entity ID: #1-#2 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  Tobacco mosaic virus (strain vulgare) Tobacco mosaic virus (strain vulgare) |
| Details of virus | Empty: NO / Enveloped: NO / Isolate: STRAIN / Type: VIRION |
| Natural host | Organism: Nicotiana tabacum |
| Buffer solution | pH: 7 / Details: pure water |
| Specimen | Conc.: 22 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: Pelco Easyglow, factory settings / Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: C-flat-2/2 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 283.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 215000 X / Nominal defocus max: 350 nm / Nominal defocus min: 150 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 5 sec. / Electron dose: 30.8 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 62 |
| EM imaging optics | Energyfilter name: GIF Quantum LS / Energyfilter slit width: 20 eV |
| Image scans | Movie frames/image: 20 / Used frames/image: 1-20 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Helical symmerty | Angular rotation/subunit: 22.038 ° / Axial rise/subunit: 1.406 Å / Axial symmetry: C1 | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 21709 | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 1.9 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 21598 / Algorithm: BACK PROJECTION / Symmetry type: HELICAL | ||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 4UDV Accession code: 4UDV / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller










 PDBj
PDBj




