+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Mycobacterium tuberculosis encapsulin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Encapsulin / icosahedral / oxidative stress response / STRUCTURAL PROTEIN | |||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / extracellular region / plasma membrane / Type 1 encapsulin shell protein Function and homology information Function and homology information | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
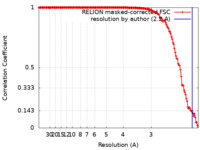
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Woodward JD | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Mycobacterium tuberculosis encapsulin Authors: Woodward JD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13420.map.gz emd_13420.map.gz | 288.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13420-v30.xml emd-13420-v30.xml emd-13420.xml emd-13420.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13420_fsc.xml emd_13420_fsc.xml | 15.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13420.png emd_13420.png | 209.9 KB | ||
| Filedesc metadata |  emd-13420.cif.gz emd-13420.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13420 http://ftp.pdbj.org/pub/emdb/structures/EMD-13420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13420 | HTTPS FTP |
-Related structure data
| Related structure data |  7phmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13420.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13420.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
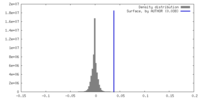
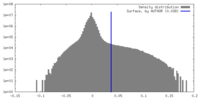
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Empty encapsulin nanocompartment
| Entire | Name: Empty encapsulin nanocompartment |
|---|---|
| Components |
|
-Supramolecule #1: Empty encapsulin nanocompartment
| Supramolecule | Name: Empty encapsulin nanocompartment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 1.73 MDa |
-Macromolecule #1: 29 kDa antigen CFP29
| Macromolecule | Name: 29 kDa antigen CFP29 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 29.554072 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNNLYRDLAP VTEAAWAEIE LEAARTFKRH IAGRRVVDVS DPGGPVTAAV STGRLIDVKA PTNGVIAHLR ASKPLVRLRV PFTLSRNEI DDVERGSKDS DWEPVKEAAK KLAFVEDRTI FEGYSAASIE GIRSASSNPA LTLPEDPREI PDVISQALSE L RLAGVDGP ...String: MNNLYRDLAP VTEAAWAEIE LEAARTFKRH IAGRRVVDVS DPGGPVTAAV STGRLIDVKA PTNGVIAHLR ASKPLVRLRV PFTLSRNEI DDVERGSKDS DWEPVKEAAK KLAFVEDRTI FEGYSAASIE GIRSASSNPA LTLPEDPREI PDVISQALSE L RLAGVDGP YSVLLSADVY TKVSETSDHG YPIREHLNRL VDGDIIWAPA IDGAFVLTTR GGDFDLQLGT DVAIGYASHD TD TVRLYLQ ETLTFLCYTA EASVALSHHH HHH UniProtKB: Type 1 encapsulin shell protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Eluted in: 50mM Tris-HCl, 350mM NaCl, 10mM Imidazol, 10% v/v glycerol at pH 7.4 and diluted 1:2 in distilled water. | |||||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa Details: Glow-discharged using an EMS100X in air: 25 mA at 20 Pa for 30 seconds. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | C-flat 2/2 holey carbon film supported by a standard copper TEM grid and coated with an ultrathin (2-3 nm) continuous carbon film. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 14133 / Average electron dose: 45.0 e/Å2 Details: Data collected in super-resolution mode at 0.53 A/pixel and down-sampled during motion correction to 1.06 A/pixel. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model was generated ab initio using Buccaneer within the CCPEM package and manually corrected using Coot before being refined using Isolde within ChimeraX and Phenix Real Space Refinement within the CCPEM package. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-7phm: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)