-Search query
-Search result
Showing all 43 items for (author: tejero & o)

EMDB-49252: 
In-situ structure of the flagellar motor of Campylobacter jejuni fcpMNO deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49253: 
In-situ structure of the flagellar motor of Campylobacter jejuni pflD deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49254: 
In-situ structure of the flagellar motor of Campylobacter jejuni flgY deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49255: 
In-situ structure of the flagellar motor of Campylobacter jejuni pflB deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49256: 
In-situ structure of the flagellar motor of Campylobacter jejuni pflA deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49257: 
In-situ structure of the flagellar motor of Campylobacter jejuni rpoN deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-49325: 
In-situ structure of the flagellar motor of Campylobacter jejuni pflC deletion mutant
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73072: 
Half of Campylobacter jejuni fcpMNO deletion mutant flagellar motor structure in situ
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73073: 
Half of Campylobacter jejuni pflD deletion mutant flagellar motor structure in situ
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73074: 
Half of Campylobacter jejuni motA deletion mutant flagellar motor structure in situ
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73075: 
Half of Campylobacter jejuni flagellar motor structure in situ
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73076: 
Focused refinement map of in situ E-ring structure in Campylobacter jejuni flagellar motor
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-73078: 
Focused refinement map of in situ spoke-rim structure in Campylobacter jejuni flagellar motor
Method: subtomogram averaging / : Tachiyama S, Liu J

EMDB-45507: 
In situ structure of C. jejuni flagellar motor
Method: subtomogram averaging / : Tachiyama S, Zhao H, Liu J

EMDB-45508: 
In situ structure of Campylobacter jejuni flagellar motor from flgX deletion mutant
Method: subtomogram averaging / : Tachiyama S, Zhao H, Liu J

EMDB-45509: 
In situ structure of C. jejuni flagellar motor from motA deletion mutant
Method: subtomogram averaging / : Tachiyama S, Zhao H, Liu J

EMDB-43188: 
Salmonella effector protein SipA decorated actin filament
Method: helical / : Guo EZ, Galan JE

PDB-8vfm: 
Salmonella effector protein SipA decorated actin filament
Method: helical / : Guo EZ, Galan JE

EMDB-17343: 
Cryo-EM structure of Rhodopsin-Gi bound to antibody fragment Fab13
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

EMDB-17344: 
Cryo-EM structure of Rhodopsin-Gi bound with antibody fragments scFv16 and Fab79, conformation 1
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

EMDB-17345: 
Cryo-EM structure of Rhodopsin-Gi bound with antibody fragments scFv16 and Fab79, conformation 2
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

PDB-8p12: 
Cryo-EM structure of Rhodopsin-Gi bound to antibody fragment Fab13
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

PDB-8p13: 
Cryo-EM structure of Rhodopsin-Gi bound with antibody fragments scFv16 and Fab79, conformation 1
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

PDB-8p15: 
Cryo-EM structure of Rhodopsin-Gi bound with antibody fragments scFv16 and Fab79, conformation 2
Method: single particle / : Pamula F, Tejero O, Muehle J, Thoma R, Schertler GFX, Marino J, Tsai CJ

EMDB-19884: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Gi heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

PDB-9epr: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Gi heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

EMDB-19882: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

EMDB-19883: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

PDB-9epp: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

PDB-9epq: 
Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer
Method: single particle / : Tejero O, Pamula F, Koyanagi M, Nagata T, Afanasyev P, Das I, Deupi X, Sheves M, Terakita A, Schertler GFX, Rodrigues MJ, Tsai CJ

EMDB-41046: 
Subtomogram average structure of the injectisome
Method: subtomogram averaging / : Park D

EMDB-41047: 
Intermembrane Tether between SCV and Mitochondria
Method: subtomogram averaging / : Park D, Liu J, Galan J

EMDB-20830: 
Cryo-EM reconstruction of the inner membrane rings containing the core components of the export apparatus and associated membranes of the needle complex from Salmonella typhimurium type III secretion system.
Method: single particle / : Butan C, Galan J

EMDB-20831: 
Cryo-EM reconstruction shows that the needle complex's inner rings from Salmonella assemble with 23-fold symmetry in the absence of the export apparatus.
Method: single particle / : Butan C, Galan J

EMDB-20832: 
Cryo-EM structure of the PrgHK periplasmic ring from the Salmonella SPI-1 type III secretion needle complex solved at 3.3 angstrom resolution
Method: single particle / : Butan C, Galan J

EMDB-20833: 
Cryo-EM reconstruction of the PrgHK periplasmic ring from Salmonella's needle complex assembled in the absence of the export apparatus
Method: single particle / : Butan C, Galan J

PDB-6uot: 
Cryo-EM structure of the PrgHK periplasmic ring from the Salmonella SPI-1 type III secretion needle complex solved at 3.3 angstrom resolution
Method: single particle / : Butan C, Galan J

PDB-6uov: 
Cryo-EM reconstruction of the PrgHK periplasmic ring from Salmonella's needle complex assembled in the absence of the export apparatus
Method: single particle / : Butan C, Galan J
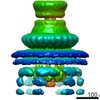
EMDB-20838: 
In situ structure of the export apparatus in Salmonella T3SS machine
Method: subtomogram averaging / : Liu J

EMDB-8545: 
SpaO mutant structure of Salmonella Type III secretion injectisome
Method: subtomogram averaging / : Hu B, Liu J

EMDB-1214: 
Assembly of the inner rod determines needle length in the type III secretion injectisome.
Method: single particle / : Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE

EMDB-1215: 
Assembly of the inner rod determines needle length in the type III secretion injectisome.
Method: single particle / : Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
