-Search query
-Search result
Showing 1 - 50 of 459 items for (author: roth & b)

EMDB-19397: 
Composite map of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS')

EMDB-19398: 
Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'')

PDB-8ro0: 
Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS')

PDB-8ro1: 
Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'')

EMDB-50447: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 1)

EMDB-50449: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 2)

EMDB-50450: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 3)

EMDB-50451: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 4)

EMDB-50452: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 5)

EMDB-50453: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 6)

EMDB-50454: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 7)

EMDB-50455: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 8)

EMDB-50456: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 9)

EMDB-50457: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 10)

EMDB-50458: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 11)

EMDB-50459: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 12)

EMDB-50460: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 13)

EMDB-50461: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 14)

EMDB-50462: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 15)

EMDB-50463: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 16)

EMDB-50464: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 17)

EMDB-50465: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 18)

EMDB-50466: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 19)

EMDB-50467: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 20)
EMDB-50468: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 21)

EMDB-50469: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 22)

EMDB-50471: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 23)

EMDB-50472: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 24)

EMDB-50473: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 25)

EMDB-50474: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 27)

EMDB-50475: 
Structure of the C. elegans Intron Lariat Spliceosome (Map 26)

PDB-9fmd: 
Integrative model of the human post-catalytic spliceosome (P-complex)

EMDB-40046: 
CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab

PDB-8ghk: 
CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab

EMDB-18290: 
Cryo-EM structure of Cx26 gap junction K125E mutant in bicarbonate buffer (classification on hemichannel)

EMDB-18291: 
Cryo-EM structure of Cx26 solubilised in LMNG - hemichannel classification - NConst conformation

EMDB-18292: 
Cryo-EM structure of Cx26 solubilised in LMNG - Hemichannel classification NFlex conformation
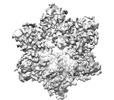
EMDB-18293: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; Nconst-mon conformation

EMDB-18294: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; NFlex conformation

EMDB-18295: 
Cryo-EM reconstruction of Cx26 gap junction K125R mutant (D6 symmetry)
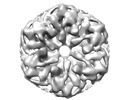
EMDB-18296: 
Cryo-EM reconstruction of Cx26 gap junction K125E mutant in HEPES buffer

EMDB-18297: 
Cryo-EM reconstruction of Cx26 gap junction WT in HEPES buffer

PDB-8q9z: 
Cryo-EM structure of Cx26 gap junction K125E mutant in bicarbonate buffer (classification on hemichannel)

PDB-8qa0: 
Cryo-EM structure of Cx26 solubilised in LMNG - hemichannel classification - NConst conformation

PDB-8qa1: 
Cryo-EM structure of Cx26 solubilised in LMNG - Hemichannel classification NFlex conformation

PDB-8qa2: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; Nconst-mon conformation

PDB-8qa3: 
Cryo-EM structure of Cx26 solubilised in LMNG: classification on subunit A; NFlex conformation

EMDB-42676: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

EMDB-42999: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

PDB-8uwl: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
