[English] 日本語
 Yorodumi
Yorodumi- EMDB-40046: CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NIAID / hemagglutinin stalk binding antibody / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
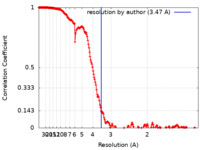
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Structural basis for the broad antigenicity of the computationally optimized influenza hemagglutinin X6. Authors: Kaito A Nagashima / John V Dzimianski / Meng Yang / Jan Abendroth / Giuseppe A Sautto / Ted M Ross / Rebecca M DuBois / Thomas E Edwards / Jarrod J Mousa /  Abstract: Influenza causes significant morbidity and mortality. As an alternative approach to current seasonal vaccines, the computationally optimized broadly reactive antigen (COBRA) platform has been ...Influenza causes significant morbidity and mortality. As an alternative approach to current seasonal vaccines, the computationally optimized broadly reactive antigen (COBRA) platform has been previously applied to hemagglutinin (HA). This approach integrates wild-type HA sequences into a single immunogen to expand the breadth of accessible antibody epitopes. Adding to previous studies of H1, H3, and H5 COBRA HAs, we define the structural features of another H1 subtype COBRA, X6, that incorporates HA sequences from before and after the 2009 H1N1 influenza pandemic. We determined structures of this antigen alone and in complex with COBRA-specific as well as broadly reactive and functional antibodies, analyzing its antigenicity. We found that X6 possesses features representing both historic and recent H1 HA strains, enabling binding to both head- and stem-reactive antibodies. Overall, these data confirm the integrity of broadly reactive antibody epitopes of X6 and contribute to design efforts for a next-generation vaccine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40046.map.gz emd_40046.map.gz | 625.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40046-v30.xml emd-40046-v30.xml emd-40046.xml emd-40046.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40046_fsc.xml emd_40046_fsc.xml | 19.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_40046.png emd_40046.png | 62.9 KB | ||
| Filedesc metadata |  emd-40046.cif.gz emd-40046.cif.gz | 6.5 KB | ||
| Others |  emd_40046_half_map_1.map.gz emd_40046_half_map_1.map.gz emd_40046_half_map_2.map.gz emd_40046_half_map_2.map.gz | 615.2 MB 615.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40046 http://ftp.pdbj.org/pub/emdb/structures/EMD-40046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40046 | HTTPS FTP |
-Related structure data
| Related structure data |  8ghkMC  8f38C  8sj9C  8v7oC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40046.map.gz / Format: CCP4 / Size: 662.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40046.map.gz / Format: CCP4 / Size: 662.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40046_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
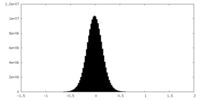
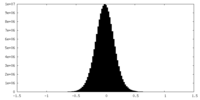
| Density Histograms |
-Half map: #1
| File | emd_40046_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
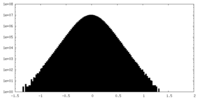
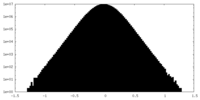
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) ...
| Entire | Name: CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) ...
| Supramolecule | Name: CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 221 KDa |
-Macromolecule #1: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: 1/Melbourne/1/1946(H1N1) Influenza A virus / Strain: 1/Melbourne/1/1946(H1N1) |
| Molecular weight | Theoretical: 53.994246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCLLKGIAPL QLGNCSVAGW ILGNPECELL ISKESWSYIV ETPNPENGT CYPGYFADYE ELREQLSSVS SFERFEIFPK ESSWPNHTVT GVSASCSHNG KSSFYRNLLW LTGKNGLYPN L SKSYANNK ...String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCLLKGIAPL QLGNCSVAGW ILGNPECELL ISKESWSYIV ETPNPENGT CYPGYFADYE ELREQLSSVS SFERFEIFPK ESSWPNHTVT GVSASCSHNG KSSFYRNLLW LTGKNGLYPN L SKSYANNK EKEVLVLWGV HHPPNIGDQR ALYHTENAYV SVVSSHYSRK FTPEIAKRPK VRDQEGRINY YWTLLEPGDT II FEANGNL IAPRYAFALS RGFGSGIITS NAPMDECDAK CQTPQGAINS SLPFQNVHPV TIGECPKYVR SAKLRMVTGL RNI PSIAGF IEGGWTGMVD GWYGYHHQNE QGSGYAADQK STQNAINGIT NKVNSVIEKM NTQFTAVGKE FNKLERRMEN LNKK VDDGF LDIWTYNAEL LVLLENERTL DFHDSNVKNL YEKVKSQLKN NAKEIGNGCF EFYHKCNNEC MESVKNGTYD YPKYS |
-Macromolecule #2: GS10-X6-BE4 Fab Heavy chain
| Macromolecule | Name: GS10-X6-BE4 Fab Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.154263 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVLGLKWVFF VVLYQGVHCE VQLVESGGGL VQPKGSLKLS CATSGFTFNT FDMHWVRQAP GKDLEWVARI RTKGNSYATY YAASVKDRI TISRDDSQSM LYLEMNSLRS EDTAMYYCVR EGGHYYGYYF DFWGQGTTLT VSS |
-Macromolecule #3: GS10-X6-BE4 Fab light chain
| Macromolecule | Name: GS10-X6-BE4 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.949619 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSAQFLGLL LLCFQGTRCE IQMTQTTSSL SASLGDRVTI SCRASQDIYN YLNWYQQQPD GAVKLLIYYT SKLHSGVPSR FSGSGSGTD YSLTITNLEQ EDIATYFCQQ GYTLPYTFGG GTKLEIK |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: LACEY | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7031 / Average exposure time: 2.4 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)