+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the C. elegans Intron Lariat Spliceosome (Map 8) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mRNA / splicing / Intorn Lariat spliceosome / ILS / pre-mRNA | |||||||||
| Biological species |  | |||||||||
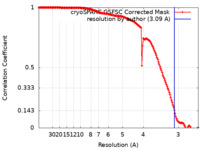
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Vorlaender MK / Rothe P / Plaschka C | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Mechanism for the initiation of spliceosome disassembly. Authors: Matthias K Vorländer / Patricia Rothe / Justus Kleifeld / Eric D Cormack / Lalitha Veleti / Daria Riabov-Bassat / Laura Fin / Alex W Phillips / Luisa Cochella / Clemens Plaschka /   Abstract: Precursor-mRNA (pre-mRNA) splicing requires the assembly, remodelling and disassembly of the multi-megadalton ribonucleoprotein complex called the spliceosome. Recent studies have shed light on ...Precursor-mRNA (pre-mRNA) splicing requires the assembly, remodelling and disassembly of the multi-megadalton ribonucleoprotein complex called the spliceosome. Recent studies have shed light on spliceosome assembly and remodelling for catalysis, but the mechanism of disassembly remains unclear. Here we report cryo-electron microscopy structures of nematode and human terminal intron lariat spliceosomes along with biochemical and genetic data. Our results uncover how four disassembly factors and the conserved RNA helicase DHX15 initiate spliceosome disassembly. The disassembly factors probe large inner and outer spliceosome surfaces to detect the release of ligated mRNA. Two of these factors, TFIP11 and C19L1, and three general spliceosome subunits, SYF1, SYF2 and SDE2, then dock and activate DHX15 on the catalytic U6 snRNA to initiate disassembly. U6 therefore controls both the start and end of pre-mRNA splicing. Taken together, our results explain the molecular basis of the initiation of canonical spliceosome disassembly and provide a framework to understand general spliceosomal RNA helicase control and the discard of aberrant spliceosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50455.map.gz emd_50455.map.gz | 324.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50455-v30.xml emd-50455-v30.xml emd-50455.xml emd-50455.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50455_fsc.xml emd_50455_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_50455.png emd_50455.png | 152.9 KB | ||
| Filedesc metadata |  emd-50455.cif.gz emd-50455.cif.gz | 4.1 KB | ||
| Others |  emd_50455_additional_1.map.gz emd_50455_additional_1.map.gz emd_50455_half_map_1.map.gz emd_50455_half_map_1.map.gz emd_50455_half_map_2.map.gz emd_50455_half_map_2.map.gz | 172.9 MB 318.8 MB 318.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50455 http://ftp.pdbj.org/pub/emdb/structures/EMD-50455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50455 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50455.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50455.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3013 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_50455_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
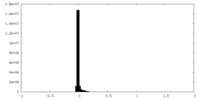
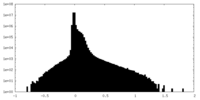
| Density Histograms |
-Half map: #1
| File | emd_50455_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
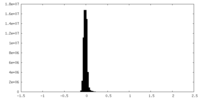
| Density Histograms |
-Half map: #2
| File | emd_50455_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
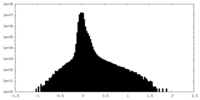
| Density Histograms |
- Sample components
Sample components
-Entire : Intron lariat spliceosome
| Entire | Name: Intron lariat spliceosome |
|---|---|
| Components |
|
-Supramolecule #1: Intron lariat spliceosome
| Supramolecule | Name: Intron lariat spliceosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#36 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Crosslinked with glutaraledhyde |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller



















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)