[English] 日本語
 Yorodumi
Yorodumi- EMDB-18291: Cryo-EM structure of Cx26 solubilised in LMNG - hemichannel class... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Cx26 solubilised in LMNG - hemichannel classification - NConst conformation | |||||||||
 Map data Map data | Full map locally sharpened in phenix; classification and masked refinement over A-F hemichannel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Gap junction Large Pore Channel Carbon dioxide sensitive / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTransport of connexons to the plasma membrane / gap junction channel activity involved in cell communication by electrical coupling / gap junction-mediated intercellular transport / Oligomerization of connexins into connexons / Transport of connexins along the secretory pathway / gap junction assembly / connexin complex / gap junction / Gap junction assembly / gap junction channel activity ...Transport of connexons to the plasma membrane / gap junction channel activity involved in cell communication by electrical coupling / gap junction-mediated intercellular transport / Oligomerization of connexins into connexons / Transport of connexins along the secretory pathway / gap junction assembly / connexin complex / gap junction / Gap junction assembly / gap junction channel activity / endoplasmic reticulum-Golgi intermediate compartment / sensory perception of sound / transmembrane transport / cell-cell signaling / calcium ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
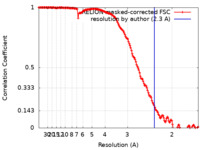
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Brotherton DH / Savva CG / Cameron AD | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structures of wild-type and a constitutively closed mutant of connexin26 shed light on channel regulation by CO. Authors: Deborah H Brotherton / Sarbjit Nijjar / Christos G Savva / Nicholas Dale / Alexander David Cameron /  Abstract: Connexins allow intercellular communication by forming gap junction channels (GJCs) between juxtaposed cells. Connexin26 (Cx26) can be regulated directly by CO. This is proposed to be mediated ...Connexins allow intercellular communication by forming gap junction channels (GJCs) between juxtaposed cells. Connexin26 (Cx26) can be regulated directly by CO. This is proposed to be mediated through carbamylation of K125. We show that mutating K125 to glutamate, mimicking the negative charge of carbamylation, causes Cx26 GJCs to be constitutively closed. Through cryo-EM we observe that the K125E mutation pushes a conformational equilibrium towards the channel having a constricted pore entrance, similar to effects seen on raising the partial pressure of CO. In previous structures of connexins, the cytoplasmic loop, important in regulation and where K125 is located, is disordered. Through further cryo-EM studies we trap distinct states of Cx26 and observe density for the cytoplasmic loop. The interplay between the position of this loop, the conformations of the transmembrane helices and the position of the N-terminal helix, which controls the aperture to the pore, provides a mechanism for regulation. #1:  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structures of wild-type and a constitutively closed mutant of connexin26 shed light on channel regulation by CO2 Authors: Brotherton DH / Nijjar S / Savva CG / Dale N / Cameron AD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18291.map.gz emd_18291.map.gz | 288.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18291-v30.xml emd-18291-v30.xml emd-18291.xml emd-18291.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18291_fsc.xml emd_18291_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_18291.png emd_18291.png | 135 KB | ||
| Masks |  emd_18291_msk_1.map emd_18291_msk_1.map | 316.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18291.cif.gz emd-18291.cif.gz | 6.7 KB | ||
| Others |  emd_18291_additional_1.map.gz emd_18291_additional_1.map.gz emd_18291_half_map_1.map.gz emd_18291_half_map_1.map.gz emd_18291_half_map_2.map.gz emd_18291_half_map_2.map.gz | 250.3 MB 250.8 MB 250.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18291 http://ftp.pdbj.org/pub/emdb/structures/EMD-18291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18291 | HTTPS FTP |
-Related structure data
| Related structure data |  8qa0MC  8q9zC  8qa1C  8qa2C  8qa3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18291.map.gz / Format: CCP4 / Size: 316.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18291.map.gz / Format: CCP4 / Size: 316.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map locally sharpened in phenix; classification and masked refinement over A-F hemichannel | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18291_msk_1.map emd_18291_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
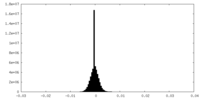
| Density Histograms |
-Additional map: Full map unsharpened; classification and masked refinement over...
| File | emd_18291_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map unsharpened; classification and masked refinement over A-F hemichannel | ||||||||||||
| Projections & Slices |
| ||||||||||||
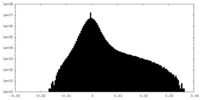
| Density Histograms |
-Half map: Half map (unsharpened); classification and masked refinement over...
| File | emd_18291_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (unsharpened); classification and masked refinement over A-F hemichannel | ||||||||||||
| Projections & Slices |
| ||||||||||||
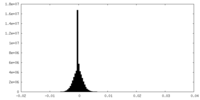
| Density Histograms |
-Half map: Half map (unsharpened); classification and masked refinement over...
| File | emd_18291_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (unsharpened); classification and masked refinement over A-F hemichannel | ||||||||||||
| Projections & Slices |
| ||||||||||||
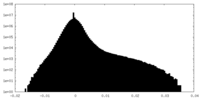
| Density Histograms |
- Sample components
Sample components
-Entire : Connexin 26, 90mmHg PCO2, pH7.4, solubilised in LMNG.
| Entire | Name: Connexin 26, 90mmHg PCO2, pH7.4, solubilised in LMNG. |
|---|---|
| Components |
|
-Supramolecule #1: Connexin 26, 90mmHg PCO2, pH7.4, solubilised in LMNG.
| Supramolecule | Name: Connexin 26, 90mmHg PCO2, pH7.4, solubilised in LMNG. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction beta-2 protein
| Macromolecule | Name: Gap junction beta-2 protein / type: protein_or_peptide / ID: 1 / Details: Connexin 26 subunit / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.713674 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDWGTLQTIL GGVNKHSTSI GKIWLTVLFI FRIMILVVAA KEVWGDEQAD FVCNTLQPGC KNVCYDHYFP ISHIRLWALQ LIFVSTPAL LVAMHVAYRR HEKKRKFIKG EIKSEFKDIE EIKTQKVRIE GSLWWTYTSS IFFRVIFEAA FMYVFYVMYD G FSMQRLVK ...String: MDWGTLQTIL GGVNKHSTSI GKIWLTVLFI FRIMILVVAA KEVWGDEQAD FVCNTLQPGC KNVCYDHYFP ISHIRLWALQ LIFVSTPAL LVAMHVAYRR HEKKRKFIKG EIKSEFKDIE EIKTQKVRIE GSLWWTYTSS IFFRVIFEAA FMYVFYVMYD G FSMQRLVK CNAWPCPNTV DCFVSRPTEK TVFTVFMIAV SGICILLNVT ELCYLLIRYC SGKSKKPVLV PR UniProtKB: Gap junction beta-2 protein |
-Macromolecule #2: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 2 / Number of copies: 24 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 342 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.7 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: The buffer, except LMNG and DTT, was prepared fresh from 10x stock on day of used. The basal buffer was filtered and degassed and DDM and DTT added. The buffer was pH corrected at point of ...Details: The buffer, except LMNG and DTT, was prepared fresh from 10x stock on day of used. The basal buffer was filtered and degassed and DDM and DTT added. The buffer was pH corrected at point of use to pH 7.4 using 15% CO2 in 85% N2. | |||||||||||||||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK I Details: Blot 3 seconds, force 10, 1 blot, skip transfer. 3ul sample, in 15% CO2/85%N2 atmosphere. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 11362 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)