-Search query
-Search result
Showing 1 - 50 of 62 items for (author: martin-benito & j)

EMDB-51384: 
Structure of Sticholisin II in large unilamellar vesicles.
Method: single particle / : Santiago C, Martin-Benito J, Arranz R, Masiulis S

EMDB-51432: 
Structure of fragacetoxin C in lipid nanodiscs
Method: single particle / : Martin Benito J, Santiago C

PDB-9gj8: 
Structure of Sticholisin II in large unilamellar vesicles.
Method: single particle / : Santiago C, Martin-Benito J, Arranz R, Masiulis S

EMDB-51426: 
Structure of the octameric pore of Fragaceotxin C (FraC or DELTA-actitoxin-Afr1a) in large unilamellar vesicles.
Method: single particle / : Martin Benito J, Santiago C, Carlero D, Arranz R

EMDB-51431: 
Structure of 6mer pore intermediate of Sticholysin II (StnII) toxin in lipid nanodiscs
Method: single particle / : Martin Benito J, Santiago C, Carlero D, Arranz R

EMDB-51420: 
Structure of 5mer pore intermediate of Sticholysin II (StnII) toxin in lipid nanodiscs
Method: single particle / : Martin Benito J, Santiago C, Carlero D

EMDB-14727: 
3D reconstruction of the cylindrical assembly of DnaJA2 by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta JM, Muga A

EMDB-14729: 
3D reconstruction of the cylindrical assembly of DnaJA2 without symmetry imposition
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta JM, Muga A

EMDB-14736: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta J, Muga A

PDB-7zhs: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Valpuesta J, Muga A

EMDB-14706: 
3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F without symmetry imposition
Method: single particle / : Cuellar J, Velasco-Carneros L, Santiago C, Martin-Benito J, Muga A, Valpuesta JM

EMDB-4423: 
Helical part of the influenza A virus ribonucleoprotein. Conformation 3.
Method: helical / : Coloma R, Arranz R

PDB-6i7b: 
Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3.
Method: helical / : Coloma R, Arranz R, de la Rosa-Trevin JM, Sorzano COS, Carlero D, Ortin J, Martin-Benito J

EMDB-0175: 
Helical part of the influenza A virus ribonucleoprotein. Conformation 1.
Method: helical / : Coloma R, Arranz R

EMDB-4426: 
Helical part of the influenza A virus ribonucleoprotein. Conformation 4.
Method: helical / : Coloma R, Arranz R

PDB-6h9g: 
Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1.
Method: helical / : Coloma R, Arranz R, de la Rosa-Trevin JM, Sorzano COS, Munier S, Carlero D, Naffakh N, Ortin J, Martin-Benito J

PDB-6i7m: 
Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4.
Method: helical / : Coloma R, Arranz R, de la Rosa-Trevin JM, Sorzano COS, Carlero D, Ortin J, Martin-Benito J

EMDB-4430: 
Helical part of the influenza A virus ribonucleoprotein. Conformation 5.
Method: helical / : Coloma R, Arranz R

PDB-6i85: 
Influenza A nucleoprotein docked into the 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 5.
Method: helical / : Coloma R, Arranz R, de la Rosa-Trevin JM, Sorzano COS, Carlero D, Ortin J, Martin-Benito J

EMDB-4412: 
Helical part of the influenza A virus ribonucleoprotein. Conformation 2.
Method: helical / : Coloma R, Arranz R

PDB-6i54: 
Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 2.
Method: helical / : Coloma R, Arranz R, de la Rosa-Trevin JM, Sorzano COS, Carlero D, Ortin J, Martin-Benito J

EMDB-0323: 
Structural insights into the ability of nucleoplasmin to assemble and chaperone histone octamers for DNA deposition
Method: single particle / : Valpuesta JM, Arranz R, Martin-Benito J

EMDB-4293: 
Hsp70:p53-TMGST:CHIP
Method: single particle / : Quintana-Gallardo L, Martin-Benito J, Valpuesta JM

EMDB-4294: 
Hsp90:p53-TMGST:CHIP
Method: single particle / : Quintana-Gallardo L, Martin-Benito J, Valpuesta JM
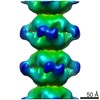
EMDB-4402: 
Electron microscopy map of human IMPDH isoform 1 bound to GDP in 150 g/L Ficoll-70
Method: helical / : Martin-Benito J, Nunez R, Fernandez-Justel D, Revuelta JL, Buey RM
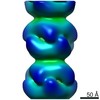
EMDB-4403: 
Electron microscopy map of human IMPDH isoform 1 bound to ATP in 150 g/L Ficoll-70
Method: helical / : Martin-Benito J, Nunez R, Fernandez-Justel D, Revuelta JL, Buey RM

EMDB-3753: 
Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium.
Method: single particle / : Scheffer MP

EMDB-3756: 
Sub-tomogram average of the surface adhesin (NAP) complex from Mycoplasma genitalium cells by cryo-electron tomography.
Method: subtomogram averaging / : Scheffer MP, Gonzalez-Gonzalez L, Frangakis AS

EMDB-2717: 
Electron cryo-microscopy of T7 bacteriophage tail after DNA ejection
Method: single particle / : Gonzalez-Garcia VA, Pulido-Cid M, Garcia-Doval C, van Raaij MJ, Martin-Benito J, Cuervo A, Carrascosa JL

EMDB-6182: 
3D structure of RepA-WH1 double filaments
Method: helical / : Torreira E, Moreno M, Fuentes-Perez ME, Fernandez C, Martin-Benito J, Moreno-Herrero F, Giraldo R, Llorca O
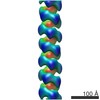
EMDB-6183: 
3D structure of RepA-WH1 single filaments
Method: helical / : Torreira E, Moreno M, Fuentes-Perez ME, Fernandez C, Martin-Benito J, Moreno-Herrero F, Giraldo R, Llorca O

EMDB-2447: 
Electron microscopy of the complex formed by chaperones TBCE and TBCB and alpha-tubulin
Method: single particle / : Serna M, Carranza G, Martin-Benito J, Janowsk R, Canals A, Coll M, Zabala JC, Valpuesta JM

EMDB-5713: 
Electron microscopy of negatively-stained gp12 tubular protein of T7 bacteriophage
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

EMDB-5689: 
Cryo-electron microscopy of T7 tail complex
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

EMDB-5690: 
Cryo-electron microscopy of T7 tail complex formed by gp8, gp11, and gp12 proteins
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

PDB-3j4a: 
Structure of gp8 connector protein
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

PDB-3j4b: 
Structure of T7 gatekeeper protein (gp11)
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL
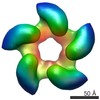
EMDB-2355: 
Negative staining three dimensional reconstruction of bacteriophage T7 large terminase
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

EMDB-2356: 
Negative staining three-dimensional reconstruction of bacteriophage T7 connector/terminase complex
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

PDB-4bij: 
Threading model of T7 large terminase
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

PDB-4bil: 
Threading model of the T7 large terminase within the gp8gp19 complex
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

EMDB-2205: 
Cryo-electron microscopy reconstruction of the helical part of influenza A virus ribonucleoprotein isolated from virions.
Method: helical / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

EMDB-2206: 
Negative stained electron microscopy reconstruction of the loop of influenza A virus ribonucleoprotein isolated from virions.
Method: single particle / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

EMDB-2207: 
Negative stained electron microscopy reconstruction of the viral polymerase of influenza A virus ribonucleoprotein isolated from virions. Conformation 1.
Method: single particle / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

EMDB-2208: 
Negative stained electron microscopy reconstruction of the viral polymerase of influenza A virus ribonucleoprotein isolated from virions. Conformation 2.
Method: single particle / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

PDB-4bbl: 
Cryo-electron microscopy reconstruction of the helical part of influenza A virus ribonucleoprotein isolated from virions.
Method: helical / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

EMDB-5505: 
Hexameric structure of the conjugative VirB4 ATPase TrwK: new insights into a functional and phylogenetic relationship with DNA translocases
Method: single particle / : Pena A, Matilla I, Martin-Benito J, Valpuesta JM, Carrascosa JL, De la Cruz F, Cabezon E, Arechaga I

EMDB-1774: 
Understanding Ribosome Assembly: The Structure of in vivo Assembled Immature 30S Subunits Revealed by Cryo-Electron Microscopy
Method: single particle / : Jomaa A, Stewart G, Benito JM, Zielke R, Campbell T, Maddock J, Brown E, Ortega J

EMDB-1775: 
Understanding Ribosome Assembly: the Structure of in vivo Assembled Immature 30S Subunits Revealed by Cryo-electron Microscopy
Method: single particle / : Jomaa A, Stewart G, Benito JM, Zielke R, Campbell T, Maddock J, Brown E, Ortega J

PDB-2xvr: 
Phage T7 empty mature head shell
Method: single particle / : Ionel A, Velazquez-Muriel JA, Luque D, Cuervo A, Caston JR, Valpuesta JM, Martin-Benito J, Carrascosa JL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
