[English] 日本語
 Yorodumi
Yorodumi- EMDB-3753: Negative stain 3D single particle reconstruction of the isolated ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3753 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
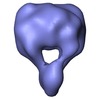
| Title | Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium. | |||||||||
 Map data Map data | Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 19.0 Å | |||||||||
 Authors Authors | Scheffer MP | |||||||||
 Citation Citation |  Journal: Mol Microbiol / Year: 2017 Journal: Mol Microbiol / Year: 2017Title: Structural characterization of the NAP; the major adhesion complex of the human pathogen Mycoplasma genitalium. Authors: Margot P Scheffer / Luis Gonzalez-Gonzalez / Anja Seybert / Mercè Ratera / Michael Kunz / José M Valpuesta / Ignacio Fita / Enrique Querol / Jaume Piñol / Jaime Martín-Benito / Achilleas S Frangakis /   Abstract: Mycoplasma genitalium, the causative agent of non-gonococcal urethritis and pelvic inflammatory disease in humans, is a small eubacterium that lacks a peptidoglycan cell wall. On the surface of its ...Mycoplasma genitalium, the causative agent of non-gonococcal urethritis and pelvic inflammatory disease in humans, is a small eubacterium that lacks a peptidoglycan cell wall. On the surface of its plasma membrane is the major surface adhesion complex, known as NAP that is essential for adhesion and gliding motility of the organism. Here, we have performed cryo-electron tomography of intact cells and detergent permeabilized M. genitalium cell aggregates, providing sub-tomogram averages of free and cell-attached NAPs respectively, revealing a tetrameric complex with two-fold rotational (C2) symmetry. Each NAP has two pairs of globular lobes (named α and β lobes), arranged as a dimer of heterodimers with each lobe connected by a stalk to the cell membrane. The β lobes are larger than the α lobes by 20%. Classification of NAPs showed that the complex can tilt with respect to the cell membrane. A protein complex containing exclusively the proteins P140 and P110, was purified from M. genitalium and was structurally characterized by negative-stain single particle EM reconstruction. The close structural similarity found between intact NAPs and the isolated P140/P110 complexes, shows that dimers of P140/P110 heterodimers are the only components of the extracellular region of intact NAPs in M. genitalium. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3753.map.gz emd_3753.map.gz | 954.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3753-v30.xml emd-3753-v30.xml emd-3753.xml emd-3753.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3753.png emd_3753.png | 78.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3753 http://ftp.pdbj.org/pub/emdb/structures/EMD-3753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3753 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3753.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3753.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative stain 3D single particle reconstruction of the isolated ...
| Entire | Name: Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium. |
|---|---|
| Components |
|
-Supramolecule #1: Negative stain 3D single particle reconstruction of the isolated ...
| Supramolecule | Name: Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium. type: complex / ID: 1 / Parent: 0 Details: NAP complexes were extracted from the cell membranes of a modified M. genitalium G37 strain by using the detergent n-octyl-beta-D-glucopyranoside, where gene mg192 had been extended to ...Details: NAP complexes were extracted from the cell membranes of a modified M. genitalium G37 strain by using the detergent n-octyl-beta-D-glucopyranoside, where gene mg192 had been extended to include a region coding for a six-histidines tag at the C-terminus end of the P110 protein, the gene product of mg192. |
|---|---|
| Source (natural) | Organism:  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) |
| Recombinant expression | Organism:  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) |
| Molecular weight | Experimental: 495 kDa/nm |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 14375 |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















