[English] 日本語
 Yorodumi
Yorodumi- EMDB-4402: Electron microscopy map of human IMPDH isoform 1 bound to GDP in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4402 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron microscopy map of human IMPDH isoform 1 bound to GDP in 150 g/L Ficoll-70 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 18.0 Å | |||||||||
 Authors Authors | Martin-Benito J / Nunez R / Fernandez-Justel D / Revuelta JL / Buey RM | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity. Authors: David Fernández-Justel / Rafael Núñez / Jaime Martín-Benito / David Jimeno / Adrián González-López / Eva María Soriano / José Luis Revuelta / Rubén M Buey /  Abstract: Inosine 5'-monophosphate dehydrogenase (IMPDH) catalyzes the rate-limiting step in the de novo GTP biosynthetic pathway and plays essential roles in cell proliferation. As a clinical target, IMPDH ...Inosine 5'-monophosphate dehydrogenase (IMPDH) catalyzes the rate-limiting step in the de novo GTP biosynthetic pathway and plays essential roles in cell proliferation. As a clinical target, IMPDH has been studied for decades, but it has only been within the last years that we are starting to understand the complexity of the mechanisms of its physiological regulation. Here, we report structural and functional insights into how adenine and guanine nucleotides control a conformational switch that modulates the assembly of the two human IMPDH enzymes into cytoophidia and allosterically regulates their catalytic activity. In vitro reconstituted micron-length cytoophidia-like structures show catalytic activity comparable to unassembled IMPDH but, in turn, are more resistant to GTP/GDP allosteric inhibition. Therefore, IMPDH cytoophidia formation facilitates the accumulation of high levels of guanine nucleotides when the cell requires it. Finally, we demonstrate that most of the IMPDH retinopathy-associated mutations abrogate GTP/GDP-induced allosteric inhibition and alter cytoophidia dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4402.map.gz emd_4402.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4402-v30.xml emd-4402-v30.xml emd-4402.xml emd-4402.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4402.png emd_4402.png | 72.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4402 http://ftp.pdbj.org/pub/emdb/structures/EMD-4402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4402 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4402 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4402.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4402.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : filaments of human IMP dehydrogenase isoform 1 in complex with GDP
| Entire | Name: filaments of human IMP dehydrogenase isoform 1 in complex with GDP |
|---|---|
| Components |
|
-Supramolecule #1: filaments of human IMP dehydrogenase isoform 1 in complex with GDP
| Supramolecule | Name: filaments of human IMP dehydrogenase isoform 1 in complex with GDP type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The filaments are formed in the presence of macromolecular crowding conditions (150 g/L Ficoll-70) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Macromolecule #1: human IMP dehydrogenase isoform 1
| Macromolecule | Name: human IMP dehydrogenase isoform 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: IMP dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: gshmadylis ggtgyvpedg ltaqqlfasa dgltyndfli lpgfidfiad evdltsaltr kitlktplis spmdtvtead maiamalmgg igfihhnctp efqanevrkv kkfeqgfitd pvvlspshtv gdvleakmrh gfsgipitet gtmgsklvgi vtsrdidfla ...String: gshmadylis ggtgyvpedg ltaqqlfasa dgltyndfli lpgfidfiad evdltsaltr kitlktplis spmdtvtead maiamalmgg igfihhnctp efqanevrkv kkfeqgfitd pvvlspshtv gdvleakmrh gfsgipitet gtmgsklvgi vtsrdidfla ekdhttllse vmtprielvv apagvtlkea neilqrskkg klpivndcde lvaiiartdl kknrdyplas kdsqkqllcg aavgtreddk yrldlltqag vdvivldssq gnsvyqiamv hyikqkyphl qviggnvvta aqaknlidag vdglrvgmgc gsicitqevm acgrpqgtav ykvaeyarrf gvpiiadggi qtvghvvkal algastvmmg sllaatteap geyffsdgvr lkkyrgmgsl damekssssq kryfsegdkv kiaqgvsgsi qdkgsiqkfv pyliagiqhg cqdigarsls vlrsmmysge lkfekrtmsa qieggvhglh syekrly |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 15.6 µm / Number grids imaged: 1 / Number real images: 75 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 54929 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: JEOL |
 Movie
Movie Controller
Controller


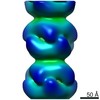
 UCSF Chimera
UCSF Chimera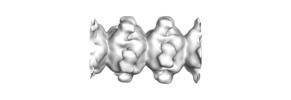







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















