-Search query
-Search result
Showing 1 - 50 of 274 items for (author: liu & cc)

EMDB-36466: 
FCP trimer in diatom Thalassiosira pseudonana

EMDB-41816: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state

EMDB-41817: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state

EMDB-41818: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state

PDB-8u1l: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state

PDB-8u1m: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state

PDB-8u1n: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state

EMDB-16933: 
Structure of cGAS in complex with SPSB3-ELOBC

EMDB-16936: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)

EMDB-16937: 
Consensus refinement of cGAS/spsb3/EloBC/Nucleosome
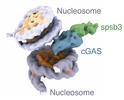
EMDB-16938: 
human cGAS/spsb3/EloBC in complex with nucleosome (2:2)

PDB-8okx: 
Structure of cGAS in complex with SPSB3-ELOBC

PDB-8ol1: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)

EMDB-26583: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

EMDB-41140: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD

EMDB-41142: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB

PDB-8tar: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD

PDB-8tau: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB

EMDB-34228: 
SARS-COV-2 BA.1 Spike incomplex with VacBB-665
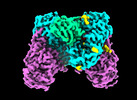
EMDB-40047: 
Structure of human ENPP1 in complex with variable heavy domain VH27.2

EMDB-33883: 
Structural basis of human PRPS2 filaments

EMDB-17154: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (consensus and constituent map 1)

EMDB-17155: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)

EMDB-17156: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 2)

EMDB-17157: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)

EMDB-17158: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (constituent map 2 from additional focus classification on PAS domains)

EMDB-17159: 
Cryo-EM map of MYC-MAX-OCT4-LIN28 complex

EMDB-17160: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)

EMDB-17161: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 1)

EMDB-17162: 
MAX-MAX bound to a nucleosome at SHL+5.1 and SHL-6.9.

EMDB-17183: 
OCT4 and MYC-MAX co-bound to a nucleosome

EMDB-17184: 
MYC-MAX bound to a nucleosome at SHL+5.8

PDB-8osj: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)

PDB-8osk: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)

PDB-8osl: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)

PDB-8ots: 
OCT4 and MYC-MAX co-bound to a nucleosome

PDB-8ott: 
MYC-MAX bound to a nucleosome at SHL+5.8

EMDB-34226: 
SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551

EMDB-34227: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 1

EMDB-34229: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 4

EMDB-34230: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 2

EMDB-34235: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 3

EMDB-34236: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 5
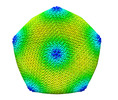
EMDB-34251: 
Singapore Grouper Iridovirus

EMDB-34815: 
One asymmetric unit of Singapore grouper iridovirus capsid

EMDB-14922: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map

PDB-7zrv: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map

EMDB-14930: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local

EMDB-14947: 
cryo-EM structure of D614 spike in complex with de novo designed binder, full and local maps(addition)

PDB-7zsd: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
