[English] 日本語
 Yorodumi
Yorodumi- EMDB-16936: cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cGAS / degradation / UPS / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology information2',3'-cyclic GMP-AMP synthase activity / cyclic GMP-AMP synthase / paracrine signaling / STING mediated induction of host immune responses / poly-ADP-D-ribose modification-dependent protein binding / target-directed miRNA degradation / elongin complex / regulation of immunoglobulin production / cGAS/STING signaling pathway / regulation of T cell activation ...2',3'-cyclic GMP-AMP synthase activity / cyclic GMP-AMP synthase / paracrine signaling / STING mediated induction of host immune responses / poly-ADP-D-ribose modification-dependent protein binding / target-directed miRNA degradation / elongin complex / regulation of immunoglobulin production / cGAS/STING signaling pathway / regulation of T cell activation / pattern recognition receptor signaling pathway / VCB complex / : / Cul5-RING ubiquitin ligase complex / negative regulation of epithelial to mesenchymal transition / SCF ubiquitin ligase complex / Cul2-RING ubiquitin ligase complex / STAT family protein binding / cytoplasmic pattern recognition receptor signaling pathway / negative regulation of cGAS/STING signaling pathway / cellular response to exogenous dsRNA / RSV-host interactions / ubiquitin-like protein ligase binding / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / positive regulation of type I interferon production / : / Tat-mediated elongation of the HIV-1 transcript / negative regulation of double-strand break repair via homologous recombination / Formation of HIV-1 elongation complex containing HIV-1 Tat / ubiquitin-like ligase-substrate adaptor activity / Formation of HIV elongation complex in the absence of HIV Tat / protein K48-linked ubiquitination / nucleosome binding / negative regulation of megakaryocyte differentiation / protein localization to CENP-A containing chromatin / RNA Polymerase II Transcription Elongation / Chromatin modifying enzymes / Formation of RNA Pol II elongation complex / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / positive regulation of defense response to virus by host / phosphatidylinositol-4,5-bisphosphate binding / Packaging Of Telomere Ends / RNA Polymerase II Pre-transcription Events / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / telomere organization / activation of innate immune response / Interleukin-7 signaling / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / transcription corepressor binding / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / determination of adult lifespan / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / TP53 Regulates Transcription of DNA Repair Genes / HDACs deacetylate histones / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / RNA Polymerase I Promoter Escape / Nonhomologous End-Joining (NHEJ) / molecular condensate scaffold activity / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / HDMs demethylate histones / Vif-mediated degradation of APOBEC3G / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / Inactivation of CSF3 (G-CSF) signaling / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / Evasion by RSV of host interferon responses / DNA Damage/Telomere Stress Induced Senescence / Pre-NOTCH Transcription and Translation / Meiotic recombination / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Regulation of expression of SLITs and ROBOs / Metalloprotease DUBs / RMTs methylate histone arginines Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Xu PB / Ablasser A | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation. Authors: Pengbiao Xu / Ying Liu / Chong Liu / Baptiste Guey / Lingyun Li / Pauline Melenec / Jonathan Ricci / Andrea Ablasser /   Abstract: Cyclic GMP-AMP synthase (cGAS) senses aberrant DNA during infection, cancer and inflammatory disease, and initiates potent innate immune responses through the synthesis of 2'3'-cyclic GMP-AMP (cGAMP). ...Cyclic GMP-AMP synthase (cGAS) senses aberrant DNA during infection, cancer and inflammatory disease, and initiates potent innate immune responses through the synthesis of 2'3'-cyclic GMP-AMP (cGAMP). The indiscriminate activity of cGAS towards DNA demands tight regulatory mechanisms that are necessary to maintain cell and tissue homeostasis under normal conditions. Inside the cell nucleus, anchoring to nucleosomes and competition with chromatin architectural proteins jointly prohibit cGAS activation by genomic DNA. However, the fate of nuclear cGAS and its role in cell physiology remains unclear. Here we show that the ubiquitin proteasomal system (UPS) degrades nuclear cGAS in cycling cells. We identify SPSB3 as the cGAS-targeting substrate receptor that associates with the cullin-RING ubiquitin ligase 5 (CRL5) complex to ligate ubiquitin onto nuclear cGAS. A cryo-electron microscopy structure of nucleosome-bound cGAS in a complex with SPSB3 reveals a highly conserved Asn-Asn (NN) minimal degron motif at the C terminus of cGAS that directs SPSB3 recruitment, ubiquitylation and cGAS protein stability. Interference with SPSB3-regulated nuclear cGAS degradation primes cells for type I interferon signalling, conferring heightened protection against infection by DNA viruses. Our research defines protein degradation as a determinant of cGAS regulation in the nucleus and provides structural insights into an element of cGAS that is amenable to therapeutic exploitation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16936.map.gz emd_16936.map.gz | 43 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16936-v30.xml emd-16936-v30.xml emd-16936.xml emd-16936.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16936.png emd_16936.png | 61.3 KB | ||
| Filedesc metadata |  emd-16936.cif.gz emd-16936.cif.gz | 7.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16936 http://ftp.pdbj.org/pub/emdb/structures/EMD-16936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16936 | HTTPS FTP |
-Related structure data
| Related structure data |  8ol1MC  8okxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16936.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16936.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.268 Å | ||||||||||||||||||||||||||||||||||||
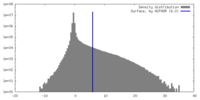
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : cGAS-Spsb3-EloBC complex
+Supramolecule #1: cGAS-Spsb3-EloBC complex
+Macromolecule #1: Histone H3.2
+Macromolecule #2: Histone H4
+Macromolecule #3: Histone H2A type 1-H
+Macromolecule #4: Histone H2B type 1-H
+Macromolecule #5: Histone H2A type 1-J
+Macromolecule #6: Histone H2B type 1-N
+Macromolecule #9: Cyclic GMP-AMP synthase
+Macromolecule #10: SPRY domain-containing SOCS box protein 3
+Macromolecule #11: Elongin-C
+Macromolecule #12: Elongin-B
+Macromolecule #7: DNA (145-MER)
+Macromolecule #8: DNA (145-MER)
+Macromolecule #13: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS buffer |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)