-Search query
-Search result
Showing 1 - 50 of 278 items for (author: kim & sk)
EMDB-41874: 
CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06

EMDB-42676: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

EMDB-42999: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

PDB-8uwl: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

PDB-8v6u: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-41248: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

EMDB-41249: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

PDB-8th3: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

PDB-8th4: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane
EMDB-15898: 
Recombinant Mayaro virus-like particle

EMDB-18941: 
SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules)

EMDB-40088: 
HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab

PDB-8gje: 
HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask

EMDB-40181: 
Human TRPV3 tetramer structure, closed conformation

EMDB-40183: 
Human TRPV3 pentamer structure

PDB-8gka: 
Human TRPV3 tetramer structure, closed conformation

PDB-8gkg: 
Human TRPV3 pentamer structure

EMDB-40062: 
Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc

EMDB-40063: 
Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc

PDB-8gi8: 
Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc

PDB-8gi9: 
Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc

EMDB-40803: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP2

EMDB-40804: 
BG505 Boost2 SOSIP.664 in complex with NHP Polyclonal Antibody FP4

EMDB-40805: 
BG505 Boost 2 in complex with NHP antibody V1V2

EMDB-40806: 
BG505 Boost 2 in complex with NHP Polyclonal Antibody N625

EMDB-40807: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base1

EMDB-40808: 
Boost 2 SOSIP.664 in complex with NHP polyclonal antibody Base2

EMDB-40809: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base3

EMDB-40810: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1

PDB-8sw7: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1

EMDB-34813: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril

EMDB-41182: 
Cryo-EM map of the Unmodified nucleosome core particle in 100 mM KCl with local resolution values
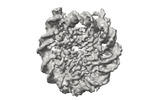
EMDB-41183: 
Cryo-EM map of the PARylated nucleosome core particle in 100 mM KCl with local resolution values
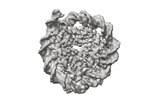
EMDB-41184: 
Cryo-EM map of the Unmodified nucleosome core particle in 5 mM KCl with local resolution values

EMDB-28228: 
SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment

PDB-8elj: 
SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment
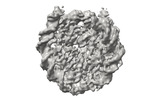
EMDB-41178: 
Cryo-EM map of the PARylated nucleosome core particle in 5 mM KCl with local resolution values
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
