[English] 日本語
 Yorodumi
Yorodumi- EMDB-41249: Structure of AT118-L Nanobody Antagonist in Complex with the Angi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan | ||||||||||||||||||||||||
 Map data Map data | Locally Refined Sharpened Map of AT118-L, AT1R-BRIL and BAG2 Anti-BRIL Fab Variable Domains in Complex with Losartan | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | G protein-coupled receptor / nanobody / SIGNALING PROTEIN-IMMUNE SYSTEM complex | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationangiotensin type I receptor activity / angiotensin type II receptor activity / phospholipase C-activating angiotensin-activated signaling pathway / regulation of renal sodium excretion / maintenance of blood vessel diameter homeostasis by renin-angiotensin / bradykinin receptor binding / renin-angiotensin regulation of aldosterone production / positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of cholesterol metabolic process / low-density lipoprotein particle remodeling ...angiotensin type I receptor activity / angiotensin type II receptor activity / phospholipase C-activating angiotensin-activated signaling pathway / regulation of renal sodium excretion / maintenance of blood vessel diameter homeostasis by renin-angiotensin / bradykinin receptor binding / renin-angiotensin regulation of aldosterone production / positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of cholesterol metabolic process / low-density lipoprotein particle remodeling / positive regulation of macrophage derived foam cell differentiation / regulation of systemic arterial blood pressure by renin-angiotensin / regulation of vasoconstriction / Rho protein signal transduction / Peptide ligand-binding receptors / blood vessel diameter maintenance / angiotensin-activated signaling pathway / cell chemotaxis / regulation of cell growth / kidney development / calcium-mediated signaling / electron transport chain / positive regulation of reactive oxygen species metabolic process / positive regulation of inflammatory response / Cargo recognition for clathrin-mediated endocytosis / regulation of cell population proliferation / Clathrin-mediated endocytosis / positive regulation of cytosolic calcium ion concentration / regulation of inflammatory response / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / electron transfer activity / periplasmic space / iron ion binding / G protein-coupled receptor signaling pathway / inflammatory response / protein heterodimerization activity / heme binding / symbiont entry into host cell / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||||||||
 Authors Authors | Skiba MA / Kruse AC | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Antibodies expand the scope of angiotensin receptor pharmacology. Authors: Meredith A Skiba / Sarah M Sterling / Shaun Rawson / Shuhao Zhang / Huixin Xu / Haoran Jiang / Genevieve R Nemeth / Morgan S A Gilman / Joseph D Hurley / Pengxiang Shen / Dean P Staus / ...Authors: Meredith A Skiba / Sarah M Sterling / Shaun Rawson / Shuhao Zhang / Huixin Xu / Haoran Jiang / Genevieve R Nemeth / Morgan S A Gilman / Joseph D Hurley / Pengxiang Shen / Dean P Staus / Jihee Kim / Conor McMahon / Maria K Lehtinen / Howard A Rockman / Patrick Barth / Laura M Wingler / Andrew C Kruse /   Abstract: G-protein-coupled receptors (GPCRs) are key regulators of human physiology and are the targets of many small-molecule research compounds and therapeutic drugs. While most of these ligands bind to ...G-protein-coupled receptors (GPCRs) are key regulators of human physiology and are the targets of many small-molecule research compounds and therapeutic drugs. While most of these ligands bind to their target GPCR with high affinity, selectivity is often limited at the receptor, tissue and cellular levels. Antibodies have the potential to address these limitations but their properties as GPCR ligands remain poorly characterized. Here, using protein engineering, pharmacological assays and structural studies, we develop maternally selective heavy-chain-only antibody ('nanobody') antagonists against the angiotensin II type I receptor and uncover the unusual molecular basis of their receptor antagonism. We further show that our nanobodies can simultaneously bind to angiotensin II type I receptor with specific small-molecule antagonists and demonstrate that ligand selectivity can be readily tuned. Our work illustrates that antibody fragments can exhibit rich and evolvable pharmacology, attesting to their potential as next-generation GPCR modulators. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41249.map.gz emd_41249.map.gz | 157 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41249-v30.xml emd-41249-v30.xml emd-41249.xml emd-41249.xml | 28.8 KB 28.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41249.png emd_41249.png | 44 KB | ||
| Filedesc metadata |  emd-41249.cif.gz emd-41249.cif.gz | 7.1 KB | ||
| Others |  emd_41249_additional_1.map.gz emd_41249_additional_1.map.gz emd_41249_additional_2.map.gz emd_41249_additional_2.map.gz emd_41249_additional_3.map.gz emd_41249_additional_3.map.gz emd_41249_half_map_1.map.gz emd_41249_half_map_1.map.gz emd_41249_half_map_2.map.gz emd_41249_half_map_2.map.gz | 154.3 MB 154.3 MB 157 MB 154.2 MB 154.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41249 http://ftp.pdbj.org/pub/emdb/structures/EMD-41249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41249 | HTTPS FTP |
-Related structure data
| Related structure data |  8th4MC  8th3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41249.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41249.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally Refined Sharpened Map of AT118-L, AT1R-BRIL and BAG2 Anti-BRIL Fab Variable Domains in Complex with Losartan | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Half Map B of Global Map of AT118-L,...
| File | emd_41249_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B of Global Map of AT118-L, AT1R-BRIL,BAG2 Anti-BRIL Fab, and anti-Fab Nanobody in Complex with Losartan | ||||||||||||
| Projections & Slices |
| ||||||||||||
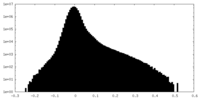
| Density Histograms |
-Additional map: Half Map A of Global Map of AT118-L,...
| File | emd_41249_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A of Global Map of AT118-L, AT1R-BRIL,BAG2 Anti-BRIL Fab, and anti-Fab Nanobody in Complex with Losartan | ||||||||||||
| Projections & Slices |
| ||||||||||||
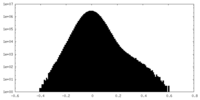
| Density Histograms |
-Additional map: Global Sharpened Map of AT118-L, AT1R-BRIL,BAG2 Anti-BRIL Fab,...
| File | emd_41249_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global Sharpened Map of AT118-L, AT1R-BRIL,BAG2 Anti-BRIL Fab, and anti-Fab Nanobody in Complex with Losartan | ||||||||||||
| Projections & Slices |
| ||||||||||||
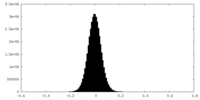
| Density Histograms |
-Half map: Half Map B of Locally Refined Sharpened Map...
| File | emd_41249_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B of Locally Refined Sharpened Map of AT118-L, AT1R-BRIL and BAG2 Anti-BRIL Fab Variable Domains in Complex with Losartan | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A of Locally Refined Sharpened Map...
| File | emd_41249_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A of Locally Refined Sharpened Map of AT118-L, AT1R-BRIL and BAG2 Anti-BRIL Fab Variable Domains in Complex with Losartan | ||||||||||||
| Projections & Slices |
| ||||||||||||
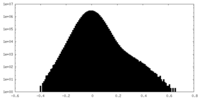
| Density Histograms |
- Sample components
Sample components
-Entire : AT118-L Nanobody Antagonist in Complex with the Angiotensin II Ty...
| Entire | Name: AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor BRIL Fusion protein, Anti-BRIL Fab, Anti-Fab Nanobody, and Losartan |
|---|---|
| Components |
|
-Supramolecule #1: AT118-L Nanobody Antagonist in Complex with the Angiotensin II Ty...
| Supramolecule | Name: AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor BRIL Fusion protein, Anti-BRIL Fab, Anti-Fab Nanobody, and Losartan type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Type-1 angiotensin II receptor Soluble cytochrome b562 complex
| Macromolecule | Name: Type-1 angiotensin II receptor Soluble cytochrome b562 complex type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.501059 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKIL NSSTEDGIKR IQDDCPKAGR HNYIFVMIPT LYSIIFVVGI FGNSLVVIVI YFYMKLKTVA SVFLLNLALA DLCFLLTLP LWAVYTAMEY RWPFGNYLCK IASASVSFNL YASVFLLTCL SIDRYLAIVH PMKSRLRRTM LVAKVTCIII W LLAGLASL ...String: DYKDDDDKIL NSSTEDGIKR IQDDCPKAGR HNYIFVMIPT LYSIIFVVGI FGNSLVVIVI YFYMKLKTVA SVFLLNLALA DLCFLLTLP LWAVYTAMEY RWPFGNYLCK IASASVSFNL YASVFLLTCL SIDRYLAIVH PMKSRLRRTM LVAKVTCIII W LLAGLASL PAIIHRNVFF IENTNITVCA FHYESQNSTL PIGLGLTKNI LGFLFPFLII LTSYTLIWKA LKKAYDLEDN WE TLNDNLK VIEKADNAAQ VKDALTKMRA AALDAQKATP PKLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQ AAAEQL KTTRNAYIQK YLERARSTLD KLNDDIFKII MAIVLFFFFS WIPHQIFTFL DVLIQLGIIR DCRIADIVDT AMPI TICIA YFNNCLNPLF YGFLGKKFKR YFLQLLKY UniProtKB: Type-1 angiotensin II receptor, Soluble cytochrome b562, Type-1 angiotensin II receptor |
-Macromolecule #2: BAG2 Anti-BRIL Fab Heavy Chain
| Macromolecule | Name: BAG2 Anti-BRIL Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 24.539314 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SCD |
-Macromolecule #3: BAG2 Anti-BRIL Fab Light Chain
| Macromolecule | Name: BAG2 Anti-BRIL Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.541164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QYLYYSLVTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: AT118-L Nanobody
| Macromolecule | Name: AT118-L Nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.345063 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGYIYS RYRMGWYRQA PGKGREFVAA ISGGSSTNYA DSVKGRFTIS RDNSKNTVYL QMNSLRAED TAVYYCAAYK IDSNPRVYWG QGTQVTVSSG KPIPNPLLGL DSTLEHHHHH H |
-Macromolecule #5: [2-butyl-5-chloranyl-3-[[4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]ph...
| Macromolecule | Name: [2-butyl-5-chloranyl-3-[[4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]imidazol-4-yl]methanol type: ligand / ID: 5 / Number of copies: 1 / Formula: LSN |
|---|---|
| Molecular weight | Theoretical: 422.911 Da |
| Chemical component information |  ChemComp-LSN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20 mM HEPES, pH 7.4, 100 mM NaCl, 0.05% GDN, 0.005% CHS |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Software | Name: SerialEM (ver. 4.0.5) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)