[English] 日本語
 Yorodumi
Yorodumi- EMDB-41182: Cryo-EM map of the Unmodified nucleosome core particle in 100 mM ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the Unmodified nucleosome core particle in 100 mM KCl with local resolution values | |||||||||||||||
 Map data Map data | Unsharpened map of unmodified nucleosome core particle, 100 mM KCl | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Nucleosome core particle / local resolution / PARylation / PARP / NUCLEAR PROTEIN | |||||||||||||||
| Biological species |  | |||||||||||||||
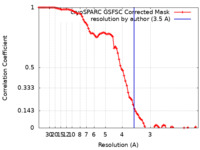
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Huang SK / Kay LE / Rubinstein JL | |||||||||||||||
| Funding support |  Canada, 4 items Canada, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Poly(ADP-ribosyl)ation enhances nucleosome dynamics and organizes DNA damage repair components within biomolecular condensates. Authors: Michael L Nosella / Tae Hun Kim / Shuya Kate Huang / Robert W Harkness / Monica Goncalves / Alisia Pan / Maria Tereshchenko / Siavash Vahidi / John L Rubinstein / Hyun O Lee / Julie D Forman-Kay / Lewis E Kay /  Abstract: Nucleosomes, the basic structural units of chromatin, hinder recruitment and activity of various DNA repair proteins, necessitating modifications that enhance DNA accessibility. Poly(ADP-ribosyl) ...Nucleosomes, the basic structural units of chromatin, hinder recruitment and activity of various DNA repair proteins, necessitating modifications that enhance DNA accessibility. Poly(ADP-ribosyl)ation (PARylation) of proteins near damage sites is an essential initiation step in several DNA-repair pathways; however, its effects on nucleosome structural dynamics and organization are unclear. Using NMR, cryoelectron microscopy (cryo-EM), and biochemical assays, we show that PARylation enhances motions of the histone H3 tail and DNA, leaving the configuration of the core intact while also stimulating nuclease digestion and ligation of nicked nucleosomal DNA by LIG3. PARylation disrupted interactions between nucleosomes, preventing self-association. Addition of LIG3 and XRCC1 to PARylated nucleosomes generated condensates that selectively partition DNA repair-associated proteins in a PAR- and phosphorylation-dependent manner in vitro. Our results establish that PARylation influences nucleosomes across different length scales, extending from the atom-level motions of histone tails to the mesoscale formation of condensates with selective compositions. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41182.map.gz emd_41182.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41182-v30.xml emd-41182-v30.xml emd-41182.xml emd-41182.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41182_fsc.xml emd_41182_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41182.png emd_41182.png | 67.2 KB | ||
| Filedesc metadata |  emd-41182.cif.gz emd-41182.cif.gz | 4 KB | ||
| Others |  emd_41182_additional_1.map.gz emd_41182_additional_1.map.gz emd_41182_additional_2.map.gz emd_41182_additional_2.map.gz emd_41182_half_map_1.map.gz emd_41182_half_map_1.map.gz emd_41182_half_map_2.map.gz emd_41182_half_map_2.map.gz | 59.7 MB 2.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41182 http://ftp.pdbj.org/pub/emdb/structures/EMD-41182 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41182 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41182 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41182.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41182.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of unmodified nucleosome core particle, 100 mM KCl | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map of unmodified nucleosome core particle, 100 mM KCl
| File | emd_41182_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of unmodified nucleosome core particle, 100 mM KCl | ||||||||||||
| Projections & Slices |
| ||||||||||||
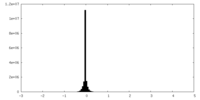
| Density Histograms |
-Additional map: Local resolution map for the map of unmodified...
| File | emd_41182_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution map for the map of unmodified nucleosome core particle, 100 mM KCl | ||||||||||||
| Projections & Slices |
| ||||||||||||
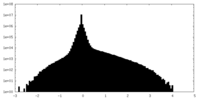
| Density Histograms |
-Half map: Half map (A) of unmodified nucleosome core particle, 100 mM KCl
| File | emd_41182_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (A) of unmodified nucleosome core particle, 100 mM KCl | ||||||||||||
| Projections & Slices |
| ||||||||||||
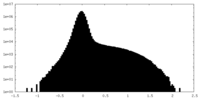
| Density Histograms |
-Half map: Half map (B) of unmodified nucleosome core particle, 100 mM KCl
| File | emd_41182_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (B) of unmodified nucleosome core particle, 100 mM KCl | ||||||||||||
| Projections & Slices |
| ||||||||||||
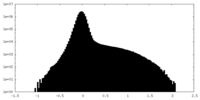
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleosome core particle
| Entire | Name: Nucleosome core particle |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome core particle
| Supramolecule | Name: Nucleosome core particle / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)