-Search query
-Search result
Showing all 46 items for (author: huang & wy)

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
Method: single particle / : Hsu HF, Wu MH, Chang YC, Hsu STD

PDB-8xbf: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
Method: single particle / : Hsu HF, Wu MH, Chang YC, Hsu STD

EMDB-29659: 
CryoEM structure of nuclear GAPDH under 8h Oxidative Stress
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-29660: 
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-29661: 
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress, class2
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-29662: 
CryoEM structure of nuclear GAPDH under 24h Oxidative Stress
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-29663: 
CryoEM structure of cytoplasmic GAPDH under 24h Oxidative Stress
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-29664: 
CryoEM structure of wild-type GAPDH
Method: single particle / : Choi WY, Wu H, Cheng YF, Manglik A

EMDB-34055: 
histone methyltransferase
Method: single particle / : Li H, Wang WY
EMDB-34053: 
histone methyltransferase
Method: single particle / : Li H, Wang WY

EMDB-34056: 
histone methyltransferase
Method: single particle / : Li H, Wang WY
EMDB-34057: 
histone methyltransferase
Method: single particle / : Li H, Wang WY

EMDB-25419: 
Previously uncharacterized rectangular bacteria in the dolphin mouth
Method: electron tomography / : Dudek NK, Galaz-Montoya JG, Shi H, Mayer M, Danita C, Celis AI, Wu GH, Behr B, Huang KC, Chiu W, Relman DA

EMDB-33149: 
SSV19
Method: single particle / : Liu HR, Chen WY

EMDB-33148: 
Tail structure of bacteriophage SSV19
Method: single particle / : Liu HR, Chen WY
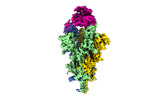
EMDB-32856: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C2 state
Method: single particle / : Han WY, Wang YF
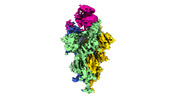
EMDB-32857: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C3 state
Method: single particle / : Han WY, Wang YF

EMDB-32558: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein with ACE2, C1 state
Method: single particle / : Han WY, Wang YF

EMDB-32559: 
Cryo-EM structure of Omicron S-ACE2, C2 state
Method: single particle / : Han WY, Wang YF

EMDB-32560: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein with human ACE2 (focus refinement on RBD-1/ACE2)
Method: single particle / : Han WY, Wang YF

EMDB-32557: 
SARS-CoV-2 Omicron S-open
Method: single particle / : Li JW, Cong Y

EMDB-30134: 
GP8 of Mature Bacteriophage T7
Method: single particle / : Chen WY, Xiao H

EMDB-30135: 
Mature bacteriorphage t7 tail adaptor protein gp11
Method: single particle / : Chen WY, Xiao H

EMDB-30136: 
Mature bacteriorphage t7 tail nozzle protein gp12
Method: single particle / : Chen WY, Xiao H

EMDB-30137: 
N-teminal of mature bacteriophage T7 tail fiber protein gp17
Method: single particle / : Chen WY, Xiao H

EMDB-30138: 
Packing Bacteriophage T7 portal protein GP8
Method: single particle / : Chen WY, Xiao H

EMDB-30139: 
Mature Phage Capsid
Method: single particle / : Chen WY

EMDB-30140: 
Mature Phage T7 mismatch
Method: single particle / : Chen WY, Xiao H

EMDB-30141: 
Mature Phage t7 core
Method: single particle / : Chen WY, Xiao H

EMDB-30142: 
phage t7 capsid-II
Method: single particle / : Chen WY, Xiao H

EMDB-30144: 
Phage t7 cpasid-II mismatch
Method: single particle / : Chen WY, Xiao H

EMDB-23160: 
Prefusion-stabilized SARS-CoV-2 spike bound by the engineered human antibody ADG-2
Method: single particle / : Wrapp D, McLellan JS

EMDB-30145: 
Phage t7 capsid-II core
Method: single particle / : Chen WY, Xiao H

EMDB-22137: 
Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies
Method: single particle / : Franklin MC, Saotome K, Romero Hernandez A, Zhou Y

EMDB-20293: 
EBOV GPdTM (Mayinga) in complex with rEBOV-548 Fab
Method: single particle / : Ward AB, Turner HL, Murin CD

EMDB-20301: 
EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs
Method: single particle / : Ward AB, Murin CD, Alkutkar T

EMDB-20947: 
EBOV GPdMuc Makona bound to rEBOV-548 Fab
Method: single particle / : Murin CD, Ward AB

PDB-6pci: 
EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs
Method: single particle / : Ward AB, Murin CD, Alkutkar T

EMDB-7955: 
Negative stain EM map of Pan-EBOV antibody 520 Fab in complex with EBOV GPdMuc trimer
Method: single particle / : Ward AB, Turner HL

EMDB-7956: 
Negative stain EM map of Pan-EBOV antibody 515 Fab in complex with EBOV GPdMuc trimer
Method: single particle / : Ward AB, Turner HL

EMDB-6640: 
cryo-EM map of the full-length human NPC1 at 4.4 angstrom
Method: single particle / : Gong X, Qiang HW, Zhou XH, Wu JP, Zhou Q, Yan N

EMDB-6641: 
cryo-EM map of the full-length human NPC1 at 6.7 angstrom
Method: single particle / : Gong X, Qiang HW, Zhou XH, Wu JP, Zhou Q, Yan N

EMDB-6480: 
Atomic structure of the Apaf-1 apoptosome
Method: single particle / : Zhou MY, Li YN, Hu Q, Bai XC, Huang WY, Yan CY, Scheres SHW, Shi YG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
