+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
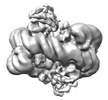
| Title | histone methyltransferase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | histone methyltransferase / GENE REGULATION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Li H / Wang WY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1. Authors: Li Huang / Youwang Wang / Haizhen Long / Haoqiang Zhu / Zengqi Wen / Liwei Zhang / Wenhao Zhang / Zhenqian Guo / Longge Wang / Fangyi Tang / Jie Hu / Keyan Bao / Ping Zhu / Guohong Li / Zheng Zhou /  Abstract: DNA replication ensures the accurate transmission of genetic information during the cell cycle. Histone variant H2A.Z is crucial for early replication origins licensing and activation in which ...DNA replication ensures the accurate transmission of genetic information during the cell cycle. Histone variant H2A.Z is crucial for early replication origins licensing and activation in which SUV420H1 preferentially recognizes H2A.Z-nucleosome and deposits H4 lysine 20 dimethylation (H4K20me2) on replication origins. Here, we report the cryo-EM structures of SUV420H1 bound to H2A.Z-nucleosome or H2A-nucleosome and demonstrate that SUV420H1 directly interacts with H4 N-terminal tail, the DNA, and the acidic patch in the nucleosome. The H4 (1-24) forms a lasso-shaped structure that stabilizes the SUV420H1-nucleosome complex and precisely projects the H4K20 residue into the SUV420H1 catalytic center. In vitro and in vivo analyses reveal a crucial role of the SUV420H1 KR loop (residues 214-223), which lies close to the H2A.Z-specific residues D97/S98, in H2A.Z-nucleosome preferential recognition. Together, our findings elucidate how SUV420H1 recognizes nucleosomes to ensure site-specific H4K20me2 modification and provide insights into how SUV420H1 preferentially recognizes H2A.Z nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34057.map.gz emd_34057.map.gz | 26.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34057-v30.xml emd-34057-v30.xml emd-34057.xml emd-34057.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34057.png emd_34057.png | 79 KB | ||
| Others |  emd_34057_half_map_1.map.gz emd_34057_half_map_1.map.gz emd_34057_half_map_2.map.gz emd_34057_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34057 http://ftp.pdbj.org/pub/emdb/structures/EMD-34057 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34057 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34057 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34057.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34057.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34057_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34057_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : histone methyltransferase and nucleosome complex
| Entire | Name: histone methyltransferase and nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: histone methyltransferase and nucleosome complex
| Supramolecule | Name: histone methyltransferase and nucleosome complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#9 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 22500 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)