+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2672 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 35O22 Fab in complex with BG505 SOSIP.664 Trimer | |||||||||
 Map data Map data | Negative stain reconstruction of 35O22 Fab bound to BG505 SOSIP trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / Env / glycoprotein / bnAb / antibody / Fab | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 19.0 Å | |||||||||
 Authors Authors | Lee JH / Pancera M / Kwong PD / Connors M / Ward AB | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Authors: Jinghe Huang / Byong H Kang / Marie Pancera / Jeong Hyun Lee / Tommy Tong / Yu Feng / Hiromi Imamichi / Ivelin S Georgiev / Gwo-Yu Chuang / Aliaksandr Druz / Nicole A Doria-Rose / Leo Laub / ...Authors: Jinghe Huang / Byong H Kang / Marie Pancera / Jeong Hyun Lee / Tommy Tong / Yu Feng / Hiromi Imamichi / Ivelin S Georgiev / Gwo-Yu Chuang / Aliaksandr Druz / Nicole A Doria-Rose / Leo Laub / Kwinten Sliepen / Marit J van Gils / Alba Torrents de la Peña / Ronald Derking / Per-Johan Klasse / Stephen A Migueles / Robert T Bailer / Munir Alam / Pavel Pugach / Barton F Haynes / Richard T Wyatt / Rogier W Sanders / James M Binley / Andrew B Ward / John R Mascola / Peter D Kwong / Mark Connors /   Abstract: The isolation of human monoclonal antibodies is providing important insights into the specificities that underlie broad neutralization of HIV-1 (reviewed in ref. 1). Here we report a broad and ...The isolation of human monoclonal antibodies is providing important insights into the specificities that underlie broad neutralization of HIV-1 (reviewed in ref. 1). Here we report a broad and extremely potent HIV-specific monoclonal antibody, termed 35O22, which binds a novel HIV-1 envelope glycoprotein (Env) epitope. 35O22 neutralized 62% of 181 pseudoviruses with a half-maximum inhibitory concentration (IC50) <50 μg ml(-1). The median IC50 of neutralized viruses was 0.033 μg ml(-1), among the most potent thus far described. 35O22 did not bind monomeric forms of Env tested, but did bind the trimeric BG505 SOSIP.664. Mutagenesis and a reconstruction by negative-stain electron microscopy of the Fab in complex with trimer revealed that it bound to a conserved epitope, which stretched across gp120 and gp41. The specificity of 35O22 represents a novel site of vulnerability on HIV Env, which serum analysis indicates to be commonly elicited by natural infection. Binding to this new site of vulnerability may thus be an important complement to current monoclonal-antibody-based approaches to immunotherapies, prophylaxis and vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2672.map.gz emd_2672.map.gz | 13.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2672-v30.xml emd-2672-v30.xml emd-2672.xml emd-2672.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2672.png emd_2672.png | 83.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2672 http://ftp.pdbj.org/pub/emdb/structures/EMD-2672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2672 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2672.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2672.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
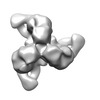
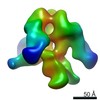
| Annotation | Negative stain reconstruction of 35O22 Fab bound to BG505 SOSIP trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
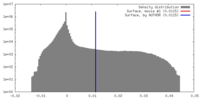
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab fragment of bnAb 35O22 bound to recombinant gp140 Env trimer ...
| Entire | Name: Fab fragment of bnAb 35O22 bound to recombinant gp140 Env trimer BG505 SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1000: Fab fragment of bnAb 35O22 bound to recombinant gp140 Env trimer ...
| Supramolecule | Name: Fab fragment of bnAb 35O22 bound to recombinant gp140 Env trimer BG505 SOSIP.664 type: sample / ID: 1000 / Oligomeric state: One Fab binds per gp140 monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Soluble HIV-1 Envelope glycoprotein
| Macromolecule | Name: Soluble HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: BG505 SOSIP.664 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / synonym: HIV-1 Human immunodeficiency virus 1 / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
-Macromolecule #2: Fab fragment of bnAb 35O22
| Macromolecule | Name: Fab fragment of bnAb 35O22 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T Homo sapiens (human) / Recombinant cell: HEK293T |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: 3 uL of protein applied, blotted then applied 2% UF for 45 seconds. |
| Grid | Details: 400 Cu mesh grid with carbon support, plasma cleaned. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TECNAI 12 |
| Temperature | Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism corrected at 52,000x mag. |
| Date | Nov 27, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 139 / Average electron dose: 30 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | FEI TECNAI 12 |
| Temperature | Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism corrected at 52,000x mag. |
| Date | Jan 22, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 139 / Average electron dose: 30 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: OTHER / Software - Name: EMAN2, SPARX / Number images used: 4746 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: H / Chain - #1 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Three copies of the Fab were fit into the EM map |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: E / Chain - #3 - Chain ID: F / Chain - #4 - Chain ID: I / Chain - #5 - Chain ID: J |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)