+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30138 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Packing Bacteriophage T7 portal protein GP8 | |||||||||
 Map data Map data | Packing Phage GP8 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | packing / phage / portal / protein / STRUCTURAL PROTEIN | |||||||||
| Function / homology | Portal protein, Caudovirales / Head-to-tail connector protein, podovirus-type / Bacteriophage head to tail connecting protein / viral portal complex / symbiont genome ejection through host cell envelope, short tail mechanism / viral DNA genome packaging / Portal protein Function and homology information Function and homology information | |||||||||
| Biological species |   Escherichia phage T7 (virus) Escherichia phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Chen WY / Xiao H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2020 Journal: Protein Cell / Year: 2020Title: Structural changes of a bacteriophage upon DNA packaging and maturation. Authors: Wenyuan Chen / Hao Xiao / Xurong Wang / Shuanglin Song / Zhen Han / Xiaowu Li / Fan Yang / Li Wang / Jingdong Song / Hongrong Liu / Lingpeng Cheng /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30138.map.gz emd_30138.map.gz | 115.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30138-v30.xml emd-30138-v30.xml emd-30138.xml emd-30138.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30138.png emd_30138.png | 42.7 KB | ||
| Filedesc metadata |  emd-30138.cif.gz emd-30138.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30138 http://ftp.pdbj.org/pub/emdb/structures/EMD-30138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30138 | HTTPS FTP |
-Related structure data
| Related structure data |  7bp0MC  7bouC  7boxC  7boyC  7bozC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30138.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30138.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Packing Phage GP8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
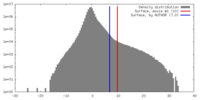
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Packing Bacteriophage T7 portal protein GP8
| Entire | Name: Packing Bacteriophage T7 portal protein GP8 |
|---|---|
| Components |
|
-Supramolecule #1: Packing Bacteriophage T7 portal protein GP8
| Supramolecule | Name: Packing Bacteriophage T7 portal protein GP8 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 59.173984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEKRTGLAE DGAKSVYERL KNDRAPYETR AQNCAQYTIP SLFPKDSDNA STDYQTPWQA VGARGLNNLA SKLMLALFPM QTWMRLTIS EYEAKQLLSD PDGLAKVDEG LSMVERIIMN YIESNSYRVT LFEALKQLVV AGNVLLYLPE PEGSNYNPMK L YRLSSYVV ...String: MAEKRTGLAE DGAKSVYERL KNDRAPYETR AQNCAQYTIP SLFPKDSDNA STDYQTPWQA VGARGLNNLA SKLMLALFPM QTWMRLTIS EYEAKQLLSD PDGLAKVDEG LSMVERIIMN YIESNSYRVT LFEALKQLVV AGNVLLYLPE PEGSNYNPMK L YRLSSYVV QRDAFGNVLQ MVTRDQIAFG ALPEDIRKAV EGQGGEKKAD ETIDVYTHIY LDEDSGEYLR YEEVEGMEVQ GS DGTYPKE ACPYIPIRMV RLDGESYGRS YIEEYLGDLR SLENLQEAIV KMSMISSKVI GLVNPAGITQ PRRLTKAQTG DFV TGRPED ISFLQLEKQA DFTVAKAVSD AIEARLSFAF MLNSAVQRTG ERVTAEEIRY VASELEDTLG GVYSILSQEL QLPL VRVLL KQLQATQQIP ELPKEAVEPT ISTGLEAIGR GQDLDKLERC VTAWAALAPM RDDPDINLAM IKLRIANAIG IDTSG ILLT EEQKQQKMAQ QSMQMGMDNG AAALAQGMAA QATASPEAMA AAADSVGLQP GI UniProtKB: Portal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 27.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)