[English] 日本語
 Yorodumi
Yorodumi- EMDB-4667: Cryo-EM structure of T7 bacteriophage portal protein, 13mer, clos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4667 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of T7 bacteriophage portal protein, 13mer, closed valve | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology | Portal protein, Caudovirales / Head-to-tail connector protein, podovirus-type / Bacteriophage head to tail connecting protein / viral portal complex / symbiont genome ejection through host cell envelope, short tail mechanism / viral DNA genome packaging / Portal protein Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | ||||||||||||||||||||||||
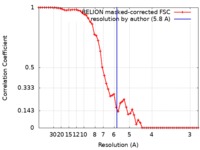
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | ||||||||||||||||||||||||
 Authors Authors | Cuervo A / Fabrega-Ferrer M / Machon C / Fernandez FJ / Perez-Luque R / Pous J / Vega MC / Carrascosa JL / Coll M | ||||||||||||||||||||||||
| Funding support |  Spain, 7 items Spain, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism. Authors: Ana Cuervo / Montserrat Fàbrega-Ferrer / Cristina Machón / José Javier Conesa / Francisco J Fernández / Rosa Pérez-Luque / Mar Pérez-Ruiz / Joan Pous / M Cristina Vega / José L Carrascosa / Miquel Coll /  Abstract: Double-stranded DNA bacteriophages package their genome at high pressure inside a procapsid through the portal, an oligomeric ring protein located at a unique capsid vertex. Once the DNA has been ...Double-stranded DNA bacteriophages package their genome at high pressure inside a procapsid through the portal, an oligomeric ring protein located at a unique capsid vertex. Once the DNA has been packaged, the tail components assemble on the portal to render the mature infective virion. The tail tightly seals the ejection conduit until infection, when its interaction with the host membrane triggers the opening of the channel and the viral genome is delivered to the host cell. Using high-resolution cryo-electron microscopy and X-ray crystallography, here we describe various structures of the T7 bacteriophage portal and fiber-less tail complex, which suggest a possible mechanism for DNA retention and ejection: a portal closed conformation temporarily retains the genome before the tail is assembled, whereas an open portal is found in the tail. Moreover, a fold including a seven-bladed β-propeller domain is described for the nozzle tail protein. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4667.map.gz emd_4667.map.gz | 14.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4667-v30.xml emd-4667-v30.xml emd-4667.xml emd-4667.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4667_fsc.xml emd_4667_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4667.png emd_4667.png | 246.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4667 http://ftp.pdbj.org/pub/emdb/structures/EMD-4667 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4667 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4667 | HTTPS FTP |
-Related structure data
| Related structure data |  4669C  4706C  6qwpC  6qx5C  6qxmC  6r21C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4667.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4667.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage packaging protein
| Entire | Name: Bacteriophage packaging protein |
|---|---|
| Components |
|
-Supramolecule #1: Bacteriophage packaging protein
| Supramolecule | Name: Bacteriophage packaging protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
| Recombinant expression | Organism:  |
| Sequence | String: MAEKRTGLAE DGAKSVYERL KNDRAPYETR AQNCAQYTIP SLFPKDSDNA STDYQTPWQA VGARGLNNL ASKLMLALFP MQTWMRLTIS EYEAKQLLSD PDGLAKVDEG LSMVERIIMN Y IESNSYRV TLFEALKQLV VAGNVLLYLP EPEGSNYNPM KLYRLSSYVV ...String: MAEKRTGLAE DGAKSVYERL KNDRAPYETR AQNCAQYTIP SLFPKDSDNA STDYQTPWQA VGARGLNNL ASKLMLALFP MQTWMRLTIS EYEAKQLLSD PDGLAKVDEG LSMVERIIMN Y IESNSYRV TLFEALKQLV VAGNVLLYLP EPEGSNYNPM KLYRLSSYVV QRDAFGNVLQ MV TRDQIAF GALPEDIRKA VEGQGGEKKA DETIDVYTHI YLDEDSGEYL RYEEVEGMEV QGS DGTYPK EACPYIPIRM VRLDGESYGR SYIEEYLGDL RSLENLQEAI VKMSMISSKV IGLV NPAGI TQPRRLTKAQ TGDFVTGRPE DISFLQLEKQ ADFTVAKAVS DAIEARLSFA FMLNS AVQR TGERVTAEEI RYVASELEDT LGGVYSILSQ ELQLPLVRVL LKQLQATQQI PELPKE AVE PTISTGLEAI GRGQDLDKLE RCVTAWAALA PMRDDPDINL AMIKLRIANA IGIDTSG IL LTEEQKQQKM AQQSMQMGMD NGAAALAQGM AAQATASPEA MAAAADSVGL QPGIAAALEH HHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 22.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)