+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30136 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mature bacteriorphage t7 tail nozzle protein gp12 | |||||||||
 Map data Map data | Mature phage GP12 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage / portal / mature / protein / STRUCTURAL PROTEIN | |||||||||
| Function / homology | virus tail, tube / symbiont genome ejection through host cell envelope, short tail mechanism / Tail tubular protein gp12 Function and homology information Function and homology information | |||||||||
| Biological species |   Escherichia phage T7 (virus) Escherichia phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Chen WY / Xiao H | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2020 Journal: Protein Cell / Year: 2020Title: Structural changes of a bacteriophage upon DNA packaging and maturation. Authors: Wenyuan Chen / Hao Xiao / Xurong Wang / Shuanglin Song / Zhen Han / Xiaowu Li / Fan Yang / Li Wang / Jingdong Song / Hongrong Liu / Lingpeng Cheng /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30136.map.gz emd_30136.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30136-v30.xml emd-30136-v30.xml emd-30136.xml emd-30136.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30136.png emd_30136.png | 105.1 KB | ||
| Filedesc metadata |  emd-30136.cif.gz emd-30136.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30136 http://ftp.pdbj.org/pub/emdb/structures/EMD-30136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30136 | HTTPS FTP |
-Related structure data
| Related structure data |  7boyMC  7bouC  7boxC  7bozC  7bp0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30136.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30136.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mature phage GP12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
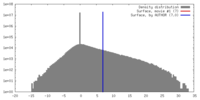
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mature bacteriorphage t7 tail nozzle protein gp12
| Entire | Name: Mature bacteriorphage t7 tail nozzle protein gp12 |
|---|---|
| Components |
|
-Supramolecule #1: Mature bacteriorphage t7 tail nozzle protein gp12
| Supramolecule | Name: Mature bacteriorphage t7 tail nozzle protein gp12 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
-Macromolecule #1: Tail tubular protein gp12
| Macromolecule | Name: Tail tubular protein gp12 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 89.480594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALISQSIKN LKGGISQQPD ILRYPDQGSR QVNGWSSETE GLQKRPPLVF LNTLGDNGAL GQAPYIHLIN RDEHEQYYAV FTGSGIRVF DLSGNEKQVR YPNGSNYIKT ANPRNDLRMV TVADYTFIVN RNVVAQKNTK SVNLPNYNPN QDGLINVRGG Q YGRELIVH ...String: MALISQSIKN LKGGISQQPD ILRYPDQGSR QVNGWSSETE GLQKRPPLVF LNTLGDNGAL GQAPYIHLIN RDEHEQYYAV FTGSGIRVF DLSGNEKQVR YPNGSNYIKT ANPRNDLRMV TVADYTFIVN RNVVAQKNTK SVNLPNYNPN QDGLINVRGG Q YGRELIVH INGKDVAKYK IPDGSQPEHV NNTDAQWLAE ELAKQMRTNL SDWTVNVGQG FIHVTAPSGQ QIDSFTTKDG YA DQLINPV THYAQSFSKL PPNAPNGYMV KIVGDASKSA DQYYVRYDAE RKVWTETLGW NTEDQVLWET MPHALVRAAD GNF DFKWLE WSPKSCGDVD TNPWPSFVGS SINDVFFFRN RLGFLSGENI ILSRTAKYFN FYPASIANLS DDDPIDVAVS TNRI AILKY AVPFSEELLI WSDEAQFVLT ASGTLTSKSV ELNLTTQFDV QDRARPFGIG RNVYFASPRS SFTSIHRYYA VQDVS SVKN AEDITSHVPN YIPNGVFSIC GSGTENFCSV LSHGDPSKIF MYKFLYLNEE LRQQSWSHWD FGENVQVLAC QSISSD MYV ILRNEFNTFL ARISFTKNAI DLQGEPYRAF MDMKIRYTIP SGTYNDDTFT TSIHIPTIYG ANFGRGKITV LEPDGKI TV FEQPTAGWNS DPWLRLSGNL EGRMVYIGFN INFVYEFSKF LIKQTADDGS TSTEDIGRLQ LRRAWVNYEN SGTFDIYV E NQSSNWKYTM AGARLGSNTL RAGRLNLGTG QYRFPVVGNA KFNTVYILSD ETTPLNIIGC GWEGNYLRRS SGI UniProtKB: Tail tubular protein gp12 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 24.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 55212 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)