-Search query
-Search result
Showing 1 - 50 of 94 items for (author: du & jj)

EMDB-37827: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state

EMDB-37828: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state

EMDB-37829: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate)

EMDB-37830: 
Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution)

EMDB-40046: 
CryoEM structure of Influenza A virus A/Melbourner/1/1946 (H1N1) hemagglutinin bound to GS10-X6-BE4 Fab

EMDB-43008: 
Fab fragment of human mAb #58 in complex with computationally optimized broadly reactive H1 influenza hemagglutinin X6

EMDB-16903: 
60S ribosomal subunit bound to the E3-UFM1 complex (native, UFM1 pulldown)

EMDB-16880: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native)

EMDB-16902: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native)

EMDB-16905: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution)

EMDB-16908: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native)

EMDB-42044: 
H2HA A/Japan/305/1957 complexed with mouse polyclonal Fab - wk 12, reoriented immunogen

EMDB-42045: 
H2HA A/Japan/305/1957 complexed with mouse polyclonal Fab - wk 12, control immunogen

EMDB-42046: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 12, control immunogen

EMDB-42048: 
H3N2 A/Victoria/3/1975 complexed with mouse polyclonal Fab - wk 7, control immunogen

EMDB-42052: 
H3N2 A/Victoria/3/1975 complexed with mouse polyclonal Fab - wk 7, reoH2HA immunogen

EMDB-42056: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, reoriented immunogen

EMDB-42058: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, control immunogen

EMDB-42059: 
H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 7, reoriented immunogen

EMDB-36057: 
Cryo-EM map of DAM (a newly designed immunogen for monkeypox virus) in complex with half of the antigen-binding fragment (Fab) of A27D7 and the single-chain fragment variable (scFv) of 7D11

EMDB-17994: 
Cryo-EM structure of a full-length HACE1 dimer

EMDB-18056: 
HACE1 in complex with RAC1 Q61L

EMDB-36056: 
Cryo-EM map of half of DAM (the A35 part) in complex with the antigen-binding fragment (Fab) of A27D7

EMDB-28833: 
Cryo-EM structure of X6 COBRA (H1N1) hemagglutinin bound to CR6261 Fab
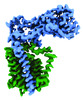
EMDB-41299: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
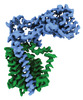
EMDB-41303: 
Transmembrane map
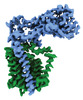
EMDB-41304: 
Periplasmic map

EMDB-16076: 
Helical shell of CCMV capsid protein on DNA origami 6HB-2k

EMDB-16077: 
Inner helical shell of CCMV capsid protein on DNA origami 6HB-10k

EMDB-16078: 
Outer helical shell of CCMV capsid protein on DNA origami 6HB-10k
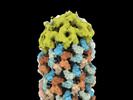
EMDB-16079: 
Helical shell cap of CCMV capsid protein on DNA origami 6HB-2k

EMDB-16080: 
Helical shell of CCMV capsid protein on DNA origami 24HB-2.5k

EMDB-29916: 
Subtomogram average of the AnaS GV shell

EMDB-26983: 
1F8 mAb in complex with the computationally optimized broadly reactive H1 influenza hemagglutinin P1

EMDB-29921: 
Subtomogram average of the native Ana GV shell

EMDB-29922: 
Cryo-tomogram of the native Ana GV

EMDB-29923: 
Cryo-tomogram of the Halo GV (c-vac)

EMDB-29924: 
Cryo-tomogram of Halo GV (p-vac)

EMDB-29925: 
Cryo-tomogram of the Mega GVs
EMDB-15205: 
cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1

EMDB-25606: 
Zika Virus particle bound with IgM antibody DH1017 Fab fragment

EMDB-32737: 
Local structure of BD55-3546 Fab and SARS-COV2 Delta RBD complex

EMDB-32331: 
Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist

EMDB-32342: 
Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist

EMDB-32732: 
Local structure of BD55-1239 Fab and SARS-COV2 Omicron RBD complex

EMDB-32738: 
Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD

EMDB-14153: 
SARS-CoV-2 Spike, C3 symmetry

EMDB-14152: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry

EMDB-14154: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C1 symmetry

EMDB-14155: 
SARS-CoV-2 Spike, C1 symmetry
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
