[English] 日本語
 Yorodumi
Yorodumi- EMDB-42046: H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | H7N9 A/Shanghai/2/2013 complexed with mouse polyclonal Fab - wk 12, control immunogen | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Fab / Hemagglutinin / polyclonal / IMMUNE SYSTEM | |||||||||
| Biological species |  H7N9 subtype (virus) / H7N9 subtype (virus) /  | |||||||||
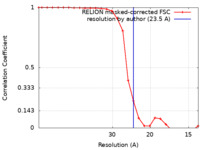
| Method | single particle reconstruction / negative staining / Resolution: 23.5 Å | |||||||||
 Authors Authors | Carter JJ / Barnes CO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Vaccine design via antigen reorientation. Authors: Duo Xu / Joshua J Carter / Chunfeng Li / Ashley Utz / Payton A B Weidenbacher / Shaogeng Tang / Mrinmoy Sanyal / Bali Pulendran / Christopher O Barnes / Peter S Kim /  Abstract: A major challenge in creating universal influenza vaccines is to focus immune responses away from the immunodominant, variable head region of hemagglutinin (HA-head) and toward the evolutionarily ...A major challenge in creating universal influenza vaccines is to focus immune responses away from the immunodominant, variable head region of hemagglutinin (HA-head) and toward the evolutionarily conserved stem region (HA-stem). Here we introduce an approach to control antigen orientation via site-specific insertion of aspartate residues that facilitates antigen binding to alum. We demonstrate the generalizability of this approach with antigens from Ebola, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza viruses and observe enhanced neutralizing antibody responses in all cases. We then reorient an H2 HA in an 'upside-down' configuration to increase the exposure and immunogenicity of HA-stem. The reoriented H2 HA (reoH2HA) on alum induced stem-directed antibodies that cross-react with both group 1 and group 2 influenza A subtypes. Electron microscopy polyclonal epitope mapping (EMPEM) revealed that reoH2HA (group 1) elicits cross-reactive antibodies targeting group 2 HA-stems. Our results highlight antigen reorientation as a generalizable approach for designing epitope-focused vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42046.map.gz emd_42046.map.gz | 434 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42046-v30.xml emd-42046-v30.xml emd-42046.xml emd-42046.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42046_fsc.xml emd_42046_fsc.xml | 2.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_42046.png emd_42046.png | 14.1 KB | ||
| Filedesc metadata |  emd-42046.cif.gz emd-42046.cif.gz | 5 KB | ||
| Others |  emd_42046_half_map_1.map.gz emd_42046_half_map_1.map.gz emd_42046_half_map_2.map.gz emd_42046_half_map_2.map.gz | 786.3 KB 786.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42046 http://ftp.pdbj.org/pub/emdb/structures/EMD-42046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42046 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42046 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42046.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42046.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42046_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
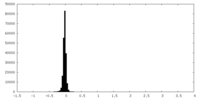
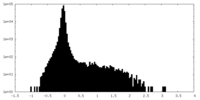
| Density Histograms |
-Half map: #2
| File | emd_42046_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
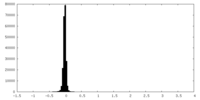
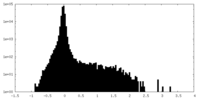
| Density Histograms |
- Sample components
Sample components
-Entire : H7HA A/Shanghai/2/2013 immune complex with mouse polyclonal Fab -...
| Entire | Name: H7HA A/Shanghai/2/2013 immune complex with mouse polyclonal Fab - week 12, control immunogen group |
|---|---|
| Components |
|
-Supramolecule #1: H7HA A/Shanghai/2/2013 immune complex with mouse polyclonal Fab -...
| Supramolecule | Name: H7HA A/Shanghai/2/2013 immune complex with mouse polyclonal Fab - week 12, control immunogen group type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinantly expressed H7HA with polyclonal Fabs generated from pooled H2HA-immunized mouse antisera |
|---|
-Supramolecule #2: H7N9 A/Shanghai/2/2013
| Supramolecule | Name: H7N9 A/Shanghai/2/2013 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  H7N9 subtype (virus) H7N9 subtype (virus) |
-Supramolecule #3: Polyclonal Fab
| Supramolecule | Name: Polyclonal Fab / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: H7N9 A/Shanghai/2/2013
| Macromolecule | Name: H7N9 A/Shanghai/2/2013 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  H7N9 subtype (virus) H7N9 subtype (virus) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNTQILVFAL IAIIPTNADK ICLGHHAVSN GTKVNTLTER GVEVVNATET VERTNIPRIC SKGKRTVDLG QCGLLGTITG PPQCDQFLEF SADLIIERRE GSDVCFPGKF VNEEALRQIL RESGGIDKEA MGFTYSGIRT NGATSACRRS GSSFYAEMKW LLSNTDNAAF ...String: MNTQILVFAL IAIIPTNADK ICLGHHAVSN GTKVNTLTER GVEVVNATET VERTNIPRIC SKGKRTVDLG QCGLLGTITG PPQCDQFLEF SADLIIERRE GSDVCFPGKF VNEEALRQIL RESGGIDKEA MGFTYSGIRT NGATSACRRS GSSFYAEMKW LLSNTDNAAF PQMTKSYKNT RKSPALIVWG IHHSVSTAEQ TKLYGSGNKL VTVGSSNYQQ SFVPSPGARP QVNGLSGRID FHWLMLNPND TVTFSFNGAF IAPDRASFLR GKSMGIQSGV QVDANCEGDC YHSGGTIISN LPFQNIDSRA VGKCPRYVKQ RSLLLATGMK NVPEIPKGRG LFGAIAGFIE NGWEGLIDGW YGFRHQNAQG EGTAADYKST QSAIDQITGK LNRLIEKTNQ QFELIDNEFN EVEKQIGNVI NWTRDSITEV WSYNAELLVA MENQHTIDLA DSEMDKLYER VKRQLRENAE EDGTGCFEIF HKCDDDCMAS IRNNTYDHSK YREEAMQNRI QIDGSMKQIE DKIEEILSKI YHIENEIARI KKLIGEVASS SGLNDIFEAQ KIEWHEAHHH HHHG |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.015 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)