-検索条件
-検索結果
検索 (著者・登録者: cianfrocco & ma)の結果52件中、1から50件目までを表示しています

EMDB-41621: 
Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)

EMDB-24672: 
The cryo-EM map of KIF18A bound to KIFBP

EMDB-24677: 
Cryo-EM structure of KIFBP core

EMDB-24744: 
Cryo-EM structure of KIFBP:KIF15

EMDB-24745: 
Cryo-EM map of KIFBP

PDB-7rsi: 
The cryo-EM map of KIF18A bound to KIFBP

PDB-7rsq: 
Cryo-EM structure of KIFBP core

PDB-7ryp: 
Cryo-EM structure of KIFBP:KIF15

PDB-7ryq: 
Cryo-EM map of KIFBP

EMDB-21692: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

EMDB-21693: 
PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma

EMDB-22754: 
Aldolase, rabbit muscle (no beam-tilt refinement)

EMDB-22755: 
Aldolase, rabbit muscle (beam-tilt refinement x1)

EMDB-22756: 
Aldolase, rabbit muscle (beam-tilt refinement x2)

EMDB-22757: 
Aldolase, rabbit muscle (beam-tilt refinement x3)

EMDB-22758: 
Aldolase, rabbit muscle (beam-tilt refinement x4)

EMDB-21490: 
Influenza hemagglutinin trimer preprocessed with automatic workflow

EMDB-21491: 
Thermoplasma acidophilum 20S proteasome cryo-EM structure using automatic preprocessing workflow

EMDB-21492: 
Rabbit muscle aldolase cryo-EM structure using automatic preprocessing workflow

EMDB-20512: 
Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome

EMDB-20513: 
Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome

EMDB-20514: 
Cryo-EM structure of RbBP5 bound to the nucleosome

EMDB-20308: 
Single Particle Reconstruction of Phosphatidylinositol (3,4,5) trisphosphate-dependent Rac exchanger 1 bound to G protein beta gamma subunits

EMDB-8908: 
Cryo-EM structure of beta-galactosidase using RELION on Amazon Web Services

PDB-6drv: 
Beta-galactosidase

EMDB-7038: 
RNA Pol II elongation complex bound to Rad26

EMDB-8736: 
Structural Basis forEukaryotic Transcription-Coupled Repair Initiation

EMDB-8885: 
RNA Pol II complex arrested by a CPD lesion

EMDB-8735: 
Ternary complex of RNA Pol II, transcription scaffold and Rad26

EMDB-8737: 
RNA pol II elongation complex

PDB-5vvr: 
Ternary complex of RNA Pol II, transcription scaffold and Rad26

PDB-5vvs: 
RNA pol II elongation complex

EMDB-8673: 
Cryo-EM structure of yeast cytoplasmic dynein-1 with Lis1 and ATP

EMDB-8706: 
Cryo-EM structure of yeast cytoplasmic dynein with Walker B mutation at AAA3 in presence of ATP-VO4

PDB-5vh9: 
Cryo-EM structure of yeast cytoplasmic dynein-1 with Lis1 and ATP

PDB-5vlj: 
Cryo-EM structure of yeast cytoplasmic dynein with Walker B mutation at AAA3 in presence of ATP-VO4
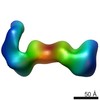
EMDB-6431: 
RCT reconstruction of CMV gHgLgO and pentameric complexes bound to highly neutralizing antibodies
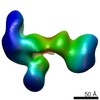
EMDB-6432: 
RCT reconstruction of glycoprotein gHgLgO in complex with neutralizing Fab fragments 3G16 and 13H11
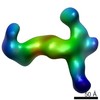
EMDB-6436: 
RCT reconstruction of CMV complex
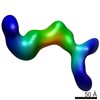
EMDB-6437: 
RCT reconstruction of CMV protein complex

EMDB-6438: 
Structure of CMV protein complex

EMDB-2858: 
Cryo-EM structure of yeast 80S ribosome calculated on Amazon's elastic cloud computing network

EMDB-2282: 
TFIIA-induced conformational changes of human TFIID suggest a modular architecture that is required for core promoter specific interactions
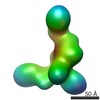
EMDB-5601: 
Substrate-specific structural rearrangements of human Dicer

EMDB-5602: 
Substrate-specific structural rearrangements of human Dicer

EMDB-5603: 
Substrate-specific structural rearrangements of human Dicer
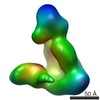
EMDB-5604: 
Substrate-specific structural rearrangements of human Dicer

EMDB-5605: 
Substrate-specific structural rearrangements of human Dicer
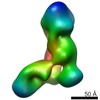
EMDB-5606: 
Substrate-specific structural rearrangements of human Dicer

EMDB-2287: 
Human TFIID binds to core promoter DNA in a reorganized structural state
ページ:
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
