[English] 日本語
 Yorodumi
Yorodumi- EMDB-2858: Cryo-EM structure of yeast 80S ribosome calculated on Amazon's el... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2858 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast 80S ribosome calculated on Amazon's elastic cloud computing network | |||||||||
 Map data Map data | 80S yeast ribosome reconstructed using Relion from published dataset EMPIAR ID 10002. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / yeast / Relion / Amazon | |||||||||
| Biological species |  | |||||||||
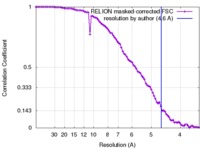
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Cianfrocco MA / Leschziner AE | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: Low cost, high performance processing of single particle cryo-electron microscopy data in the cloud. Authors: Michael A Cianfrocco / Andres E Leschziner /  Abstract: The advent of a new generation of electron microscopes and direct electron detectors has realized the potential of single particle cryo-electron microscopy (cryo-EM) as a technique to generate high- ...The advent of a new generation of electron microscopes and direct electron detectors has realized the potential of single particle cryo-electron microscopy (cryo-EM) as a technique to generate high-resolution structures. Calculating these structures requires high performance computing clusters, a resource that may be limiting to many likely cryo-EM users. To address this limitation and facilitate the spread of cryo-EM, we developed a publicly available 'off-the-shelf' computing environment on Amazon's elastic cloud computing infrastructure. This environment provides users with single particle cryo-EM software packages and the ability to create computing clusters with 16-480+ CPUs. We tested our computing environment using a publicly available 80S yeast ribosome dataset and estimate that laboratories could determine high-resolution cryo-EM structures for $50 to $1500 per structure within a timeframe comparable to local clusters. Our analysis shows that Amazon's cloud computing environment may offer a viable computing environment for cryo-EM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2858.map.gz emd_2858.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2858-v30.xml emd-2858-v30.xml emd-2858.xml emd-2858.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2858_fsc.xml emd_2858_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_2858.png emd_2858.png | 516.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2858 http://ftp.pdbj.org/pub/emdb/structures/EMD-2858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2858 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2858.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2858.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 80S yeast ribosome reconstructed using Relion from published dataset EMPIAR ID 10002. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.77 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
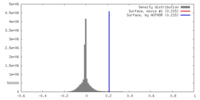
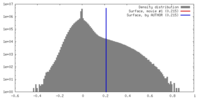
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 80S yeast ribosome
| Entire | Name: 80S yeast ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: 80S yeast ribosome
| Supramolecule | Name: 80S yeast ribosome / type: sample / ID: 1000 Details: Micrographs were downloaded from EMPIAR dataset 10002, associated with the publication Bai et al. eLife (2013) Oligomeric state: ribosome-eukaryote / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 4.2 MDa / Theoretical: 4.2 MDa |
-Supramolecule #1: 80S ribosome
| Supramolecule | Name: 80S ribosome / type: complex / ID: 1 Details: Micrographs were downloaded from EMPAIR database 10002, associated with Bai et al. eLife (2013) Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 4.2 MDa / Theoretical: 4.2 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.45 Details: 3mM Hepes-KOH, 6.6 mM Tris-acetate pH 7.2, 3 mM NH4Cl, 6.6 mM NH4-acetate, 48 mM K-acetate, 4 mM Mg-acetate, 2.4 mM DTT |
| Grid | Details: Quantifoil grids (2/2) with 3 nm thin carbon on top |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: FEI VITROBOT MARK II Details: Micrographs were downloaded from EMPIAR database 10002, associated with Bai et al. eLife (2013) Method: Blot 2.5 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 90 K / Average: 85 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 59,000 times magnification |
| Details | Micrographs were downloaded from EMPIAR 10002 |
| Date | Jul 15, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON I (4k x 4k) / Digitization - Sampling interval: 14 µm / Number real images: 200 / Average electron dose: 16 e/Å2 Details: Every image is the average of sixteen frames recorded by the direct electron detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 79096 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.44 µm / Nominal defocus min: 1.191 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID:  3u5b |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Each PDB model was docked using "Fit in map" in Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID:  3u5c |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Each PDB model was docked using "Fit in map" in Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 3
| Initial model | PDB ID:  3u5d |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Each PDB model was docked using "Fit in map" in Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 4
| Initial model | PDB ID:  3u5e |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Each PDB model was docked using "Fit in map" in Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)