+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22758 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aldolase, rabbit muscle (beam-tilt refinement x4) | |||||||||
 Map data Map data | Aldolase, rabbit muscle (beam-tilt refinement x4) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycolysis / gluconeogenesis / carbohydrate degradation / homotetramer / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of Arp2/3 complex-mediated actin nucleation / fructose-bisphosphate aldolase / fructose-bisphosphate aldolase activity / M band / I band / glycolytic process / protein homotetramerization / positive regulation of cell migration Similarity search - Function | |||||||||
| Biological species |  | |||||||||
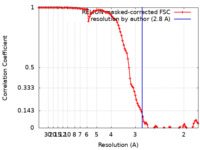
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Kearns SK / Cash JN | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2020 Journal: IUCrJ / Year: 2020Title: High-resolution cryo-EM using beam-image shift at 200 keV. Authors: Jennifer N Cash / Sarah Kearns / Yilai Li / Michael A Cianfrocco /  Abstract: Recent advances in single-particle cryo-electron microscopy (cryo-EM) data collection utilize beam-image shift to improve throughput. Despite implementation on 300 keV cryo-EM instruments, it ...Recent advances in single-particle cryo-electron microscopy (cryo-EM) data collection utilize beam-image shift to improve throughput. Despite implementation on 300 keV cryo-EM instruments, it remains unknown how well beam-image-shift data collection affects data quality on 200 keV instruments and the extent to which aberrations can be computationally corrected. To test this, a cryo-EM data set for aldolase was collected at 200 keV using beam-image shift and analyzed. This analysis shows that the instrument beam tilt and particle motion initially limited the resolution to 4.9 Å. After particle polishing and iterative rounds of aberration correction in , a 2.8 Å resolution structure could be obtained. This analysis demonstrates that software correction of microscope aberrations can provide a significant improvement in resolution at 200 keV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22758.map.gz emd_22758.map.gz | 96.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22758-v30.xml emd-22758-v30.xml emd-22758.xml emd-22758.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22758_fsc.xml emd_22758_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22758.png emd_22758.png | 89.6 KB | ||
| Filedesc metadata |  emd-22758.cif.gz emd-22758.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22758 http://ftp.pdbj.org/pub/emdb/structures/EMD-22758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22758 | HTTPS FTP |
-Related structure data
| Related structure data |  7ka4MC  7k9lC  7k9xC  7ka2C  7ka3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10519 (Title: Single particle cryo electron microscopy of aldolase (rabbit, muscle) using beam-tilt on Talos Arctica EMPIAR-10519 (Title: Single particle cryo electron microscopy of aldolase (rabbit, muscle) using beam-tilt on Talos ArcticaData size: 87.6 Data #1: Unaligned micrographs of aldolase collected with beam-tilt at 200 kV [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22758.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22758.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Aldolase, rabbit muscle (beam-tilt refinement x4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Homotetramer of aldolase
| Entire | Name: Homotetramer of aldolase |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer of aldolase
| Supramolecule | Name: Homotetramer of aldolase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 157 kDa/nm |
-Macromolecule #1: Fructose-bisphosphate aldolase A
| Macromolecule | Name: Fructose-bisphosphate aldolase A / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: fructose-bisphosphate aldolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.263672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PHSHPALTPE QKKELSDIAH RIVAPGKGIL AADESTGSIA KRLQSIGTEN TEENRRFYRQ LLLTADDRVN PCIGGVILFH ETLYQKADD GRPFPQVIKS KGGVVGIKVD KGVVPLAGTN GETTTQGLDG LSERCAQYKK DGADFAKWRC VLKIGEHTPS A LAIMENAN ...String: PHSHPALTPE QKKELSDIAH RIVAPGKGIL AADESTGSIA KRLQSIGTEN TEENRRFYRQ LLLTADDRVN PCIGGVILFH ETLYQKADD GRPFPQVIKS KGGVVGIKVD KGVVPLAGTN GETTTQGLDG LSERCAQYKK DGADFAKWRC VLKIGEHTPS A LAIMENAN VLARYASICQ QNGIVPIVEP EILPDGDHDL KRCQYVTEKV LAAVYKALSD HHIYLEGTLL KPNMVTPGHA CT QKYSHEE IAMATVTALR RTVPPAVTGV TFLSGGQSEE EASINLNAIN KCPLLKPWAL TFSYGRALQA SALKAWGGKK ENL KAAQEE YVKRALANSL ACQGKYTPSG QAGAAASESL FISNHAY UniProtKB: Fructose-bisphosphate aldolase A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | Pure aldolase isolated from rabbit muscle was purchased as a lyophilized powder (Sigma Aldrich) and solubilized in 20 mM HEPES (pH 7.5), 50 mM NaCl at 1.6 mg/ml. Sample was blotted for 4 seconds with Whatman No. #1 filter paper immediately prior to plunge freezing in liquid ethane cooled by liquid nitrogen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 4500 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.002 µm / Nominal defocus min: 0.0008 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)