+Search query
-Structure paper
| Title | Substrate-specific structural rearrangements of human Dicer. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 20, Issue 6, Page 662-670, Year 2013 |
| Publish date | Apr 28, 2013 |
 Authors Authors | David W Taylor / Enbo Ma / Hideki Shigematsu / Michael A Cianfrocco / Cameron L Noland / Kuniaki Nagayama / Eva Nogales / Jennifer A Doudna / Hong-Wei Wang /  |
| PubMed Abstract | Dicer has a central role in RNA-interference pathways by cleaving double-stranded RNAs (dsRNAs) to produce small regulatory RNAs. Human Dicer can process long double-stranded and hairpin precursor ...Dicer has a central role in RNA-interference pathways by cleaving double-stranded RNAs (dsRNAs) to produce small regulatory RNAs. Human Dicer can process long double-stranded and hairpin precursor RNAs to yield short interfering RNAs (siRNAs) and microRNAs (miRNAs), respectively. Previous studies have shown that pre-miRNAs are cleaved more rapidly than pre-siRNAs in vitro and are the predominant natural Dicer substrates. We have used EM and single-particle analysis of Dicer-RNA complexes to gain insight into the structural basis for human Dicer's substrate preference. Our studies show that Dicer traps pre-siRNAs in a nonproductive conformation, whereas interactions of Dicer with pre-miRNAs and dsRNA-binding proteins induce structural changes in the enzyme that enable productive substrate recognition in the central catalytic channel. These findings implicate RNA structure and cofactors in determining substrate recognition and processing efficiency by human Dicer. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:23624860 / PubMed:23624860 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 26.0 - 31.0 Å |
| Structure data | 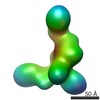 EMDB-5601:  EMDB-5602:  EMDB-5603: 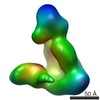 EMDB-5604:  EMDB-5605: 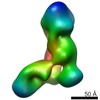 EMDB-5606: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Homo sapiens (human)
Homo sapiens (human)