-Search query
-Search result
Showing all 42 items for (author: chanda & b)

EMDB-47447: 
Glucagon Like Peptide Receptor-1 (GLP1R) A316T mutant with GLP-1 peptide. Dominant negative Gs complex.
Method: single particle / : Deane-Alder K, Belousoff MJ, Wootten DL

PDB-9e2a: 
Glucagon Like Peptide Receptor-1 (GLP1R) A316T mutant with GLP-1 peptide. Dominant negative Gs complex.
Method: single particle / : Deane-Alder K, Belousoff MJ, Wootten DL

EMDB-49659: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

EMDB-49901: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

PDB-9nqj: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

PDB-9nxc: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

EMDB-44435: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

PDB-9bcr: 
Cryo-EM structure of a bacterial prototype ATP-binding cassette transporter MalFGK2.
Method: single particle / : Qian R, Jing W, Vinay I, Shanwen Z, Jeehae S, William GL, Luis MRH, Jong HS, Young AG, IIya L, Kirill M, Baron C, Huan B

EMDB-45671: 
Subtomogram average of the Polar Tube Outer Filament layer from Encephalitozoon intestinalis microsporidian spores
Method: subtomogram averaging / : Usmani M, Coudray N, Bobe D, Kopylov M, Ekiert DC, Bhabha G

EMDB-45672: 
Subtomogram average of the Polar Tube Inner Filament layer from Encephalitozoon intestinalis microsporidian spores
Method: subtomogram averaging / : Usmani M, Coudray N, Bobe D, Kopylov M, Ekiert DC, Bhabha G

EMDB-45673: 
Subtomogram average of the Polar Tube Outer Filament Layer and Inner Filament layer from Encephalitozoon intestinalis microsporidian spores
Method: subtomogram averaging / : Usmani M, Coudray N, Bobe D, Kopylov M, Ekiert DC, Bhabha G

EMDB-45674: 
Subtomogram average of a whole Polar Tube cross-section from Encephalitozoon intestinalis microsporidian spores
Method: subtomogram averaging / : Usmani M, Coudray N, Bobe D, Kopylov M, Ekiert DC, Bhabha G

EMDB-41041: 
Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

PDB-8t50: 
Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

EMDB-41040: 
Human HCN1 F186C S264C C309A bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

PDB-8t4y: 
Human HCN1 F186C S264C C309A bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

EMDB-41036: 
Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

PDB-8t4m: 
Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL
Method: single particle / : Burtscher V, Mount J, Cowgill J, Chang Y, Bickel K, Yuan P, Chanda B

EMDB-41711: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK

PDB-8ty6: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK

EMDB-40699: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-40707: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-40708: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sq9: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sqj: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode
Method: single particle / : Small GI, Darst SA, Campbell EA

PDB-8sqk: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate
Method: single particle / : Small GI, Darst SA, Campbell EA

EMDB-16799: 
Cryo-EM structure of the NINJ1 filament
Method: helical / : Degen MD, Hiller SH, Maier TM

PDB-6r8n: 
STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A
Method: single particle / : Colletier JP, Gauto D, Estrozi L, Favier A, Effantin G, Schoehn G, Boisbouvier J, Schanda P

EMDB-4179: 
Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii
Method: single particle / : Gauto DF, Estrozi LF, Schwieters CD

PDB-6f3k: 
Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii
Method: single particle / : Gauto DF, Estrozi LF, Schwieters CD, Effantin G, Macek P, Sounier R, Kerfah R, Sivertsen AC, Colletier JP, Boisbouvier J, Schoehn G, Favier A, Schanda P
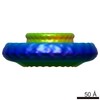
EMDB-4072: 
Cryo-EM 3D reconstruction of rings formed by the extracellular domain of SpoIIIAG from Bacillus subtilis
Method: single particle / : Rodrigues C, Henry X, Neumann E, Schoehn G, Rudner D, Morlot C

PDB-4v61: 
Homology model for the Spinach chloroplast 30S subunit fitted to 9.4A cryo-EM map of the 70S chlororibosome.
Method: single particle / : Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK

EMDB-1701: 
The molecular mechanism of the multi-tasking kinesin-8 motor
Method: helical / : Peters C, Brejc K, Belmont L, Bodey A, Lee Y, Yu M, Ramchandani S, Guo J, Lichtsteiner S, Wood KW, Sakowicz R, Hartman J, Moores C

EMDB-1702: 
Kif18A (ATP state) head bound to a microtubule
Method: helical / : Peters C, Brejc K, Belmont L, Bodey A, Lee Y, Yu M, Ramchandani S, Guo J, Lichtsteiner S, Wood KW, Sakowicz R, Hartman J, Moores C

EMDB-5125: 
PSRP1 is not a bona fide ribosomal protein, but a stress response factor
Method: single particle / : Sharma MR, Donhofer A, Barat C, Datta P, Fucini P, Wilson DN, Agrawal RK

EMDB-5126: 
In vivo 70S E.coli ribosome with PSRP1
Method: single particle / : Sharma MR, Donhofer A, Barat C, Datta P, Fucini P, Wilson DN, Agrawal RK

EMDB-1417: 
Cryo-EM study of the Spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins
Method: single particle / : Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK

EMDB-1369: 
Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits.
Method: single particle / : Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK

EMDB-1370: 
Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits.
Method: single particle / : Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK

PDB-1x18: 
Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT
Method: single particle / : Sharma MR, Barat C, Agrawal RK

PDB-1x1l: 
Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit.
Method: single particle / : Sharma MR, Barat C, Agrawal RK
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
