[English] 日本語
 Yorodumi
Yorodumi- EMDB-4179: Combined solid-state NMR, solution-state NMR and EM data for stru... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4179 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | ||||||||||||
 Map data Map data | CryoEM map of the H78C/H248C mutant tetrahedral aminopeptidase from P. horikoshii | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | peptidase / protein quality control / oligomer / aminopeptidase / PEPTIDE BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases); Aminopeptidases / aminopeptidase activity / metallopeptidase activity / proteolysis / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |   Pyrococcus horikoshii (archaea) / Pyrococcus horikoshii (archaea) /   Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3) (archaea) Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3) (archaea) | ||||||||||||
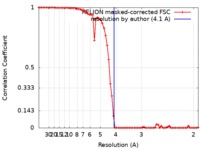
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||
 Authors Authors | Gauto DF / Estrozi LF / Schwieters CD | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex. Authors: Diego F Gauto / Leandro F Estrozi / Charles D Schwieters / Gregory Effantin / Pavel Macek / Remy Sounier / Astrid C Sivertsen / Elena Schmidt / Rime Kerfah / Guillaume Mas / Jacques-Philippe ...Authors: Diego F Gauto / Leandro F Estrozi / Charles D Schwieters / Gregory Effantin / Pavel Macek / Remy Sounier / Astrid C Sivertsen / Elena Schmidt / Rime Kerfah / Guillaume Mas / Jacques-Philippe Colletier / Peter Güntert / Adrien Favier / Guy Schoehn / Paul Schanda / Jerome Boisbouvier /      Abstract: Atomic-resolution structure determination is crucial for understanding protein function. Cryo-EM and NMR spectroscopy both provide structural information, but currently cryo-EM does not routinely ...Atomic-resolution structure determination is crucial for understanding protein function. Cryo-EM and NMR spectroscopy both provide structural information, but currently cryo-EM does not routinely give access to atomic-level structural data, and, generally, NMR structure determination is restricted to small (<30 kDa) proteins. We introduce an integrated structure determination approach that simultaneously uses NMR and EM data to overcome the limits of each of these methods. The approach enables structure determination of the 468 kDa large dodecameric aminopeptidase TET2 to a precision and accuracy below 1 Å by combining secondary-structure information obtained from near-complete magic-angle-spinning NMR assignments of the 39 kDa-large subunits, distance restraints from backbone amides and ILV methyl groups, and a 4.1 Å resolution EM map. The resulting structure exceeds current standards of NMR and EM structure determination in terms of molecular weight and precision. Importantly, the approach is successful even in cases where only medium-resolution cryo-EM data are available. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4179.map.gz emd_4179.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4179-v30.xml emd-4179-v30.xml emd-4179.xml emd-4179.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4179_fsc.xml emd_4179_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4179_1.png emd_4179_1.png emd_4179_2.png emd_4179_2.png | 111.3 KB 86.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4179 http://ftp.pdbj.org/pub/emdb/structures/EMD-4179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4179 | HTTPS FTP |
-Related structure data
| Related structure data |  6f3kMC  6r8nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4179.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4179.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of the H78C/H248C mutant tetrahedral aminopeptidase from P. horikoshii | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : tetrahedral amino-peptidase from P. horikoshii
| Entire | Name: tetrahedral amino-peptidase from P. horikoshii |
|---|---|
| Components |
|
-Supramolecule #1: tetrahedral amino-peptidase from P. horikoshii
| Supramolecule | Name: tetrahedral amino-peptidase from P. horikoshii / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: H78C/H248C mutant |
|---|---|
| Source (natural) | Organism:   Pyrococcus horikoshii (archaea) Pyrococcus horikoshii (archaea) |
| Molecular weight | Theoretical: 490 KDa |
-Macromolecule #1: Tetrahedral aminopeptidase
| Macromolecule | Name: Tetrahedral aminopeptidase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Aminopeptidases |
|---|---|
| Source (natural) | Organism:   Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3) (archaea) Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3) (archaea)Strain: ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3 |
| Molecular weight | Theoretical: 39.071027 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEVRNMVDYE LLKKVVEAPG VSGYEFLGIR DVVIEEIKDY VDEVKVDKLG NVIAHKKGEG PKVMIAAHMD QIGLMVTHIE KNGFLRVAP IGGVDPKTLI AQRFKVWIDK GKFIYGVGAS VPPHIQKPED RKKAPDWDQI FIDIGAESKE EAEDMGVKIG T VITWDGRL ...String: MEVRNMVDYE LLKKVVEAPG VSGYEFLGIR DVVIEEIKDY VDEVKVDKLG NVIAHKKGEG PKVMIAAHMD QIGLMVTHIE KNGFLRVAP IGGVDPKTLI AQRFKVWIDK GKFIYGVGAS VPPHIQKPED RKKAPDWDQI FIDIGAESKE EAEDMGVKIG T VITWDGRL ERLGKHRFVS IAFDDRIAVY TILEVAKQLK DAKADVYFVA TVQEEVGLRG ARTSAFGIEP DYGFAIDVTI AA DIPGTPE HKQVTHLGKG TAIKIMDRSV ICHPTIVRWL EELAKKHEIP YQLEILLGGG TDAGAIHLTK AGVPTGALSV PAR YIHSNT EVVDERDVDA TVELMTKALE NIHELKI UniProtKB: Tetrahedral aminopeptidase |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time 2s, force 1, drain time 0.. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 90 / Average exposure time: 4.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 25773 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 20000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-6f3k:  PDB-6r8n: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)