[English] 日本語
 Yorodumi
Yorodumi- EMDB-4072: Cryo-EM 3D reconstruction of rings formed by the extracellular do... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4072 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM 3D reconstruction of rings formed by the extracellular domain of SpoIIIAG from Bacillus subtilis | |||||||||
 Map data Map data | Cryo-EM 3D reconstruction of SpoIIIAG rings from Bacillus subtilis. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 35.0 Å | |||||||||
 Authors Authors | Rodrigues C / Henry X / Neumann E / Schoehn G / Rudner D / Morlot C | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: A ring-shaped conduit connects the mother cell and forespore during sporulation in Bacillus subtilis. Authors: Christopher D A Rodrigues / Xavier Henry / Emmanuelle Neumann / Vilius Kurauskas / Laure Bellard / Yann Fichou / Paul Schanda / Guy Schoehn / David Z Rudner / Cecile Morlot /    Abstract: During spore formation in Bacillus subtilis a transenvelope complex is assembled across the double membrane that separates the mother cell and forespore. This complex (called the "A-Q complex") is ...During spore formation in Bacillus subtilis a transenvelope complex is assembled across the double membrane that separates the mother cell and forespore. This complex (called the "A-Q complex") is required to maintain forespore development and is composed of proteins with remote homology to components of type II, III, and IV secretion systems found in Gram-negative bacteria. Here, we show that one of these proteins, SpoIIIAG, which has remote homology to ring-forming proteins found in type III secretion systems, assembles into an oligomeric ring in the periplasmic-like space between the two membranes. Three-dimensional reconstruction of images generated by cryo-electron microscopy indicates that the SpoIIIAG ring has a cup-and-saucer architecture with a 6-nm central pore. Structural modeling of SpoIIIAG generated a 24-member ring with dimensions similar to those of the EM-derived saucer. Point mutations in the predicted oligomeric interface disrupted ring formation in vitro and impaired forespore gene expression and efficient spore formation in vivo. Taken together, our data provide strong support for the model in which the A-Q transenvelope complex contains a conduit that connects the mother cell and forespore. We propose that a set of stacked rings spans the intermembrane space, as has been found for type III secretion systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4072.map.gz emd_4072.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4072-v30.xml emd-4072-v30.xml emd-4072.xml emd-4072.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4072.png emd_4072.png | 90.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4072 http://ftp.pdbj.org/pub/emdb/structures/EMD-4072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4072 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4072.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4072.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM 3D reconstruction of SpoIIIAG rings from Bacillus subtilis. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
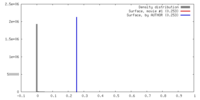
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Oligomeric rings formed by the D1+D2 extracellular domains of Spo...
| Entire | Name: Oligomeric rings formed by the D1+D2 extracellular domains of SpoIIIAG from Bacillus subtilis |
|---|---|
| Components |
|
-Supramolecule #1: Oligomeric rings formed by the D1+D2 extracellular domains of Spo...
| Supramolecule | Name: Oligomeric rings formed by the D1+D2 extracellular domains of SpoIIIAG from Bacillus subtilis type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 19 KDa |
-Macromolecule #1: SpoIIIAG
| Macromolecule | Name: SpoIIIAG / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: SSPEKTENAK TITAVSSQH SADSKEKTAE VFKASKSDKP KDSIDDYEKE YENQLKEILE TIIGVDDVSV V VNVDATSL KVYEKNKSNK NTTTEETDKE GGKRSVTDQS SEEEIVMIKN GDKETPVVVQ TK KPDIRGV LVVAQGVDNV QIKQTIIEAV TRVLDVPSHR VAVAPKKIKE DS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: KODAK SO-163 FILM / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)