[English] 日本語
 Yorodumi
Yorodumi- EMDB-41036: Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMN... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion Channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of membrane hyperpolarization / HCN channels / general adaptation syndrome, behavioral process / HCN channel complex / retinal cone cell development / intracellularly cAMP-activated cation channel activity / negative regulation of action potential / regulation of SA node cell action potential / regulation of membrane depolarization ...: / positive regulation of membrane hyperpolarization / HCN channels / general adaptation syndrome, behavioral process / HCN channel complex / retinal cone cell development / intracellularly cAMP-activated cation channel activity / negative regulation of action potential / regulation of SA node cell action potential / regulation of membrane depolarization / apical dendrite / maternal behavior / sodium ion import across plasma membrane / negative regulation of synaptic transmission, glutamatergic / response to L-glutamate / apical protein localization / voltage-gated monoatomic cation channel activity / voltage-gated sodium channel activity / potassium ion import across plasma membrane / phosphatidylinositol-3,4,5-trisphosphate binding / regulation of heart rate by cardiac conduction / voltage-gated potassium channel activity / potassium channel activity / neuronal action potential / cAMP binding / cellular response to interferon-beta / presynaptic active zone membrane / phosphatidylinositol-4,5-bisphosphate binding / potassium ion transmembrane transport / axon terminus / dendrite membrane / sodium ion transmembrane transport / cellular response to cAMP / dendritic shaft / regulation of membrane potential / response to calcium ion / protein homotetramerization / basolateral plasma membrane / postsynaptic membrane / axon / neuronal cell body / dendrite / glutamatergic synapse / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
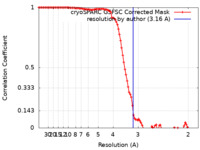
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | |||||||||
 Authors Authors | Burtscher V / Mount J / Cowgill J / Chang Y / Bickel K / Yuan P / Chanda B | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for hyperpolarization-dependent opening of human HCN1 channel. Authors: Verena Burtscher / Jonathan Mount / Jian Huang / John Cowgill / Yongchang Chang / Kathleen Bickel / Jianhan Chen / Peng Yuan / Baron Chanda /   Abstract: Hyperpolarization and cyclic nucleotide (HCN) activated ion channels are critical for the automaticity of action potentials in pacemaking and rhythmic electrical circuits in the human body. Unlike ...Hyperpolarization and cyclic nucleotide (HCN) activated ion channels are critical for the automaticity of action potentials in pacemaking and rhythmic electrical circuits in the human body. Unlike most voltage-gated ion channels, the HCN and related plant ion channels activate upon membrane hyperpolarization. Although functional studies have identified residues in the interface between the voltage-sensing and pore domain as crucial for inverted electromechanical coupling, the structural mechanisms for this unusual voltage-dependence remain unclear. Here, we present cryo-electron microscopy structures of human HCN1 corresponding to Closed, Open, and a putative Intermediate state. Our structures reveal that the downward motion of the gating charges past the charge transfer center is accompanied by concomitant unwinding of the inner end of the S4 and S5 helices, disrupting the tight gating interface observed in the Closed state structure. This helix-coil transition at the intracellular gating interface accompanies a concerted iris-like dilation of the pore helices and underlies the reversed voltage dependence of HCN channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41036.map.gz emd_41036.map.gz | 86.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41036-v30.xml emd-41036-v30.xml emd-41036.xml emd-41036.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41036_fsc.xml emd_41036_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_41036.png emd_41036.png | 143.6 KB | ||
| Filedesc metadata |  emd-41036.cif.gz emd-41036.cif.gz | 6.6 KB | ||
| Others |  emd_41036_half_map_1.map.gz emd_41036_half_map_1.map.gz emd_41036_half_map_2.map.gz emd_41036_half_map_2.map.gz | 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41036 http://ftp.pdbj.org/pub/emdb/structures/EMD-41036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41036 | HTTPS FTP |
-Validation report
| Summary document |  emd_41036_validation.pdf.gz emd_41036_validation.pdf.gz | 876 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41036_full_validation.pdf.gz emd_41036_full_validation.pdf.gz | 875.6 KB | Display | |
| Data in XML |  emd_41036_validation.xml.gz emd_41036_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_41036_validation.cif.gz emd_41036_validation.cif.gz | 23.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41036 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41036 | HTTPS FTP |
-Related structure data
| Related structure data |  8t4mMC  8t4yC  8t50C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41036.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41036.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.928 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41036_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41036_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric hHCN1
| Entire | Name: Tetrameric hHCN1 |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric hHCN1
| Supramolecule | Name: Tetrameric hHCN1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Homotetramer |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium/sodium hyperpolarization-activated cyclic nucleotide-ga...
| Macromolecule | Name: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.87182 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGGGKPNSS SNSRDDGNSV FPAKASATGA GPAAAEKRLG TPPGGGGAGA KEHGNSVCFK VDGGGGGGGG GGGGEEPAGG FEDAEGPRR QYGFMQRQFT SMLQPGVNKF SLRMFGSQKA VEKEQERVKT AGFWIIHPYS DFRFYWDLIM LIMMVGNLVI I PVGITFFT ...String: MEGGGKPNSS SNSRDDGNSV FPAKASATGA GPAAAEKRLG TPPGGGGAGA KEHGNSVCFK VDGGGGGGGG GGGGEEPAGG FEDAEGPRR QYGFMQRQFT SMLQPGVNKF SLRMFGSQKA VEKEQERVKT AGFWIIHPYS DFRFYWDLIM LIMMVGNLVI I PVGITFFT EQTTTPWIIF NVASDTVCLL DLIMNFRTGT VNEDSSEIIL DPKVIKMNYL KSWFVVDFIS SIPVDYIFLI VE KGMDSEV YKTARALRIV RFTKILCLLR LLRLSRLIRY IHQWEEIFHM TYDLASAVVR IFNLIGMMLL LCHWDGCLQF LVP LLQDFP PDCWVSLNEM VNDSWGKQYS YALFKAMSHM LCIGYGAQAP VSMSDLWITM LSMIVGATCY AMFVGHATAL IQSL DSSRR QYQEKYKQVE QYMSFHKLPA DMRQKIHDYY EHRYQGKIFD EENILNELND PLREEIVNFN CRKLVATMPL FANAD PNFV TAMLSKLRFE VFQPGDYIIR EGAVGKKMYF IQHGVAGVIT KSSKEMKLTD GSYFGEICLL TKGRRTASVR ADTYCR LYS LSVDNFNEVL EEYPMMRRAF ETVAIDRLDR IGKKNSILLQ KFQKDLNTGV FNNQENEILK QIVKHDREMV QAIAPIN YP QMTTLNSTSS TTTPTSRMRT QSPPVYTATS LSHSNLHSPS PSTQTPQPSA ILSPCSYTTA VCSPPVQSPL AARTFHYA S PTASQLSLMQ QQPQQQVQQS QPPQTQPQQP SPQPQTPGSS TPKNEVHKST QALHNTNLTR EVRPLSASQP SLPHEVSTL ISRPHPTVGE SLASIPQPVT AVPGTGLQAG GRSTVPQRVT LFRQMSSGAI PPNRGVPPAP PPPAAALPRE SSSVLNTDPD AEKPRFASN L UniProtKB: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 |
-Macromolecule #2: ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: CMP |
|---|---|
| Molecular weight | Theoretical: 329.206 Da |
| Chemical component information |  ChemComp-CMP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 1973 / Average exposure time: 9.77 sec. / Average electron dose: 53.07 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.4 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)