-Search query
-Search result
Showing 1 - 50 of 53 items for (author: bueno & t)

EMDB-19067: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

EMDB-19076: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

EMDB-19077: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).

PDB-8rd8: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

PDB-8rdv: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

PDB-8rdw: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).
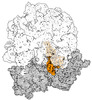
EMDB-43074: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)
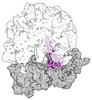
EMDB-43075: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)
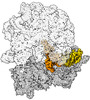
EMDB-43076: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)

EMDB-43077: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Structure 6)

EMDB-43078: 
Hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) bound to Mycobacterium smegmatis 70S ribosome, from focused 3D classification and refinement (Structure 6)

PDB-8v9j: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)

PDB-8v9k: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)

PDB-8v9l: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)

EMDB-16453: 
SARS-CoV-2 Omicron Variant Spike Trimer in complex with three 17T2 Fabs

EMDB-16473: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

PDB-8c89: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

EMDB-15243: 
Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic ecumicin (class 2)

EMDB-15240: 
Mycobacterium tuberculosis ClpC1 hexamer structure

EMDB-15241: 
Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin

EMDB-15242: 
Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1)

PDB-8a8u: 
Mycobacterium tuberculosis ClpC1 hexamer structure

PDB-8a8v: 
Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin

PDB-8a8w: 
Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1)

EMDB-13588: 
Gelsolin-free CCT

EMDB-24334: 
cryo-EM of human Gastric inhibitory polypeptide receptor GIPR bound to GIP

EMDB-24401: 
cryo-EM structure of human Gastric inhibitory polypeptide receptor GIPR bound to tirzepatide

EMDB-24445: 
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R in apo form

EMDB-24453: 
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R bound to tirzepatide

PDB-7ra3: 
cryo-EM of human Gastric inhibitory polypeptide receptor GIPR bound to GIP

PDB-7rbt: 
cryo-EM structure of human Gastric inhibitory polypeptide receptor GIPR bound to tirzepatide

PDB-7rg9: 
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R in apo form

PDB-7rgp: 
cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R bound to tirzepatide

EMDB-13177: 
Ser40 phosphorylated tyrosine hydroxylase

EMDB-13747: 
CCT-gelsolin complex

EMDB-13936: 
Cryo-EM structure of the ribosome from Encephalitozoon cuniculi

PDB-7qep: 
Cryo-EM structure of the ribosome from Encephalitozoon cuniculi

EMDB-13442: 
Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine.

PDB-7pim: 
Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine.

EMDB-11309: 
Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding.

PDB-6zn2: 
Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding.

EMDB-11624: 
Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH).

PDB-7a2g: 
Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH).

EMDB-11467: 
Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms

EMDB-11587: 
Partial structure of the substrate-free tyrosine hydroxylase (apo-TH).

PDB-6zvp: 
Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms

PDB-6zzu: 
Partial structure of the substrate-free tyrosine hydroxylase (apo-TH).

EMDB-21147: 
Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator

PDB-6vcb: 
Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator

EMDB-4489: 
Human CCT:mLST8 complex
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
