[English] 日本語
 Yorodumi
Yorodumi- EMDB-11309: Partial structure of tyrosine hydroxylase in complex with dopamin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11309 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tetramer / Dopamine / Catecholamine / Brain / Parkinson / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationtyrosine 3-monooxygenase / tyrosine 3-monooxygenase activity / dopamine biosynthetic process from tyrosine / embryonic camera-type eye morphogenesis / epinephrine biosynthetic process / Catecholamine biosynthesis / norepinephrine biosynthetic process / hyaloid vascular plexus regression / eye photoreceptor cell development / melanosome membrane ...tyrosine 3-monooxygenase / tyrosine 3-monooxygenase activity / dopamine biosynthetic process from tyrosine / embryonic camera-type eye morphogenesis / epinephrine biosynthetic process / Catecholamine biosynthesis / norepinephrine biosynthetic process / hyaloid vascular plexus regression / eye photoreceptor cell development / melanosome membrane / synaptic transmission, dopaminergic / mating behavior / eating behavior / dopamine biosynthetic process / regulation of heart contraction / pigmentation / smooth endoplasmic reticulum / anatomical structure morphogenesis / heart morphogenesis / visual perception / animal organ morphogenesis / learning / locomotory behavior / cytoplasmic side of plasma membrane / memory / synaptic vesicle / heart development / cytoplasmic vesicle / response to ethanol / perikaryon / response to hypoxia / neuron projection / iron ion binding / axon / perinuclear region of cytoplasm / enzyme binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Bueno-Carrasco MT / Cuellar J | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation. Authors: María Teresa Bueno-Carrasco / Jorge Cuéllar / Marte I Flydal / César Santiago / Trond-André Kråkenes / Rune Kleppe / José R López-Blanco / Miguel Marcilla / Knut Teigen / Sara Alvira ...Authors: María Teresa Bueno-Carrasco / Jorge Cuéllar / Marte I Flydal / César Santiago / Trond-André Kråkenes / Rune Kleppe / José R López-Blanco / Miguel Marcilla / Knut Teigen / Sara Alvira / Pablo Chacón / Aurora Martinez / José M Valpuesta /    Abstract: Tyrosine hydroxylase (TH) catalyzes the rate-limiting step in the biosynthesis of dopamine (DA) and other catecholamines, and its dysfunction leads to DA deficiency and parkinsonisms. Inhibition by ...Tyrosine hydroxylase (TH) catalyzes the rate-limiting step in the biosynthesis of dopamine (DA) and other catecholamines, and its dysfunction leads to DA deficiency and parkinsonisms. Inhibition by catecholamines and reactivation by S40 phosphorylation are key regulatory mechanisms of TH activity and conformational stability. We used Cryo-EM to determine the structures of full-length human TH without and with DA, and the structure of S40 phosphorylated TH, complemented with biophysical and biochemical characterizations and molecular dynamics simulations. TH presents a tetrameric structure with dimerized regulatory domains that are separated 15 Å from the catalytic domains. Upon DA binding, a 20-residue α-helix in the flexible N-terminal tail of the regulatory domain is fixed in the active site, blocking it, while S40-phosphorylation forces its egress. The structures reveal the molecular basis of the inhibitory and stabilizing effects of DA and its counteraction by S40-phosphorylation, key regulatory mechanisms for homeostasis of DA and TH. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11309.map.gz emd_11309.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11309-v30.xml emd-11309-v30.xml emd-11309.xml emd-11309.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11309.png emd_11309.png | 104.4 KB | ||
| Filedesc metadata |  emd-11309.cif.gz emd-11309.cif.gz | 5.5 KB | ||
| Others |  emd_11309_half_map_1.map.gz emd_11309_half_map_1.map.gz emd_11309_half_map_2.map.gz emd_11309_half_map_2.map.gz | 13.2 MB 13.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11309 http://ftp.pdbj.org/pub/emdb/structures/EMD-11309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11309 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11309 | HTTPS FTP |
-Related structure data
| Related structure data |  6zn2MC  6zvpC  6zzuC  7a2gC  7pimC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11309.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11309.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
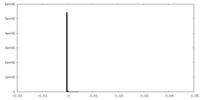
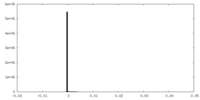
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_11309_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
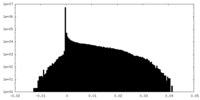
| Density Histograms |
-Half map: #1
| File | emd_11309_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
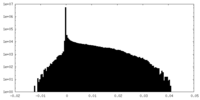
| Density Histograms |
- Sample components
Sample components
-Entire : Tyrosine Hydroxylase
| Entire | Name: Tyrosine Hydroxylase |
|---|---|
| Components |
|
-Supramolecule #1: Tyrosine Hydroxylase
| Supramolecule | Name: Tyrosine Hydroxylase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tyrosine 3-monooxygenase
| Macromolecule | Name: Tyrosine 3-monooxygenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: tyrosine 3-monooxygenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.087766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VPWFPRKVSE LDKCHHLVTK FDPDLDLDHP GFSDQVYRQR RKLIAEIAFQ YRHGDPIPRV EYTAEEIATW KEVYTTLKGL YATHACGEH LEAFALLERF SGYREDNIPQ LEDVSRFLKE RTGFQLRPVA GLLSARDFLA SLAFRVFQCT QYIRHASSPM H SPEPDCCH ...String: VPWFPRKVSE LDKCHHLVTK FDPDLDLDHP GFSDQVYRQR RKLIAEIAFQ YRHGDPIPRV EYTAEEIATW KEVYTTLKGL YATHACGEH LEAFALLERF SGYREDNIPQ LEDVSRFLKE RTGFQLRPVA GLLSARDFLA SLAFRVFQCT QYIRHASSPM H SPEPDCCH ELLGHVPMLA DRTFAQFSQD IGLASLGASD EEIEKLSTLY WFTVEFGLCK QNGEVKAYGA GLLSSYGELL HC LSEEPEI RAFDPEAAAV QPYQDQTYQS VYFVSESFSD AKDKLRSYAS RIQRPFSVKF DPYTLAIDVL DSPQAVRRSL EGV QDELDT LAHALSAIG UniProtKB: Tyrosine 3-monooxygenase |
-Macromolecule #2: SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-A...
| Macromolecule | Name: SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.874081 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLIEDARKER EAAVAAAA |
-Macromolecule #3: L-DOPAMINE
| Macromolecule | Name: L-DOPAMINE / type: ligand / ID: 3 / Number of copies: 4 / Formula: LDP |
|---|---|
| Molecular weight | Theoretical: 153.178 Da |
| Chemical component information |  ChemComp-LDP: |
-Macromolecule #4: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 125033 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)