[English] 日本語
 Yorodumi
Yorodumi- EMDB-4589: Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / lipid scrambles / TMEM16 | |||||||||
| Function / homology |  Function and homology information Function and homology informationcortical endoplasmic reticulum / chloride channel activity / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Nectria haematococca mpVI 77-13-4 (fungus) / Nectria haematococca mpVI 77-13-4 (fungus) /  Nectria haematococca (fungus) Nectria haematococca (fungus) | |||||||||
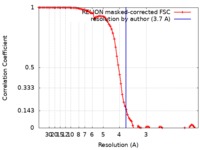
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Kalienkova V / Clerico Mosina V | |||||||||
| Funding support |  Switzerland, Switzerland,  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. Authors: Valeria Kalienkova / Vanessa Clerico Mosina / Laura Bryner / Gert T Oostergetel / Raimund Dutzler / Cristina Paulino /   Abstract: Scramblases catalyze the movement of lipids between both leaflets of a bilayer. Whereas the X-ray structure of the protein nhTMEM16 has previously revealed the architecture of a Ca-dependent lipid ...Scramblases catalyze the movement of lipids between both leaflets of a bilayer. Whereas the X-ray structure of the protein nhTMEM16 has previously revealed the architecture of a Ca-dependent lipid scramblase, its regulation mechanism has remained elusive. Here, we have used cryo-electron microscopy and functional assays to address this question. Ca-bound and Ca-free conformations of nhTMEM16 in detergent and lipid nanodiscs illustrate the interactions with its environment and they reveal the conformational changes underlying its activation. In this process, Ca binding induces a stepwise transition of the catalytic subunit cavity, converting a closed cavity that is shielded from the membrane in the absence of ligand, into a polar furrow that becomes accessible to lipid headgroups in the Ca-bound state. Additionally, our structures demonstrate how nhTMEM16 distorts the membrane at both entrances of the subunit cavity, thereby decreasing the energy barrier for lipid movement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4589.map.gz emd_4589.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4589-v30.xml emd-4589-v30.xml emd-4589.xml emd-4589.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
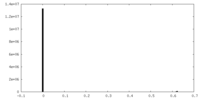
| FSC (resolution estimation) |  emd_4589_fsc.xml emd_4589_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4589.png emd_4589.png | 131.3 KB | ||
| Masks |  emd_4589_msk_1.map emd_4589_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4589.cif.gz emd-4589.cif.gz | 6.5 KB | ||
| Others |  emd_4589_additional.map.gz emd_4589_additional.map.gz emd_4589_half_map_1.map.gz emd_4589_half_map_1.map.gz emd_4589_half_map_2.map.gz emd_4589_half_map_2.map.gz | 38.9 MB 39.6 MB 39.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4589 http://ftp.pdbj.org/pub/emdb/structures/EMD-4589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4589 | HTTPS FTP |
-Related structure data
| Related structure data |  6qm6MC  4587C  4588C  4592C  4593C  4594C  6qm4C  6qm5C  6qm9C  6qmaC  6qmbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4589.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4589.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
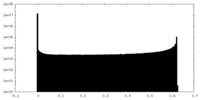
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
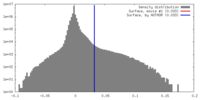
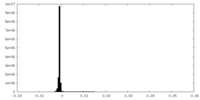
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4589_msk_1.map emd_4589_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Refined not masked nhTMEM16 DDM noCa EM map where micelle...
| File | emd_4589_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined not masked nhTMEM16_DDM_noCa EM map where micelle distortion can be observed. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 used during refinement and for...
| File | emd_4589_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 used during refinement and for FSC gold-standard resolution calculation nhTMEM16_DDM_noCa. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 used during refinement and for...
| File | emd_4589_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 used during refinement and for FSC gold-standard resolution calculation nhTMEM16_DDM_noCa. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : nhTMEM16
| Entire | Name: nhTMEM16 |
|---|---|
| Components |
|
-Supramolecule #1: nhTMEM16
| Supramolecule | Name: nhTMEM16 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Nectria haematococca mpVI 77-13-4 (fungus) Nectria haematococca mpVI 77-13-4 (fungus) |
| Molecular weight | Theoretical: 166 KDa |
-Macromolecule #1: Predicted protein
| Macromolecule | Name: Predicted protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nectria haematococca (fungus) Nectria haematococca (fungus) |
| Molecular weight | Theoretical: 83.200008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNLKDFSQP GSGQESNFGV DFVIHYKVPA AERDEAEAGF VQLIRALTTV GLATEVRHGE NESLLVFVKV ASPDLFAKQV YRARLGDWL HGVRVSAPHN DIAQALQDEP VVEAERLRLI YLMITKPHNE GGAGVTPTNA KWKHVESIFP LHSHSFNKEW I KKWSSKYT ...String: MSNLKDFSQP GSGQESNFGV DFVIHYKVPA AERDEAEAGF VQLIRALTTV GLATEVRHGE NESLLVFVKV ASPDLFAKQV YRARLGDWL HGVRVSAPHN DIAQALQDEP VVEAERLRLI YLMITKPHNE GGAGVTPTNA KWKHVESIFP LHSHSFNKEW I KKWSSKYT LEQTDIDNIR DKFGESVAFY FAFLRSYFRF LVIPSAFGFG AWLLLGQFSY LYALLCGLWS VVFFEYWKKQ EV DLAVQWG VRGVSSIQQS RPEFEWEHEA EDPITGEPVK VYPPMKRVKT QLLQIPFALA CVVALGALIV TCNSLEVFIN EVY SGPGKQ YLGFLPTIFL VIGTPTISGV LMGAAEKLNA MENYATVDAH DAALIQKQFV LNFMTSYMAL FFTAFVYIPF GHIL HPFLN FWRATAQTLT FSEKELPTRE FQINPARISN QMFYFTVTAQ IVNFATEVVV PYIKQQAFQK AKQLKSGSKV QEDHE EEAE FLQRVREECT LEEYDVSGDY REMVMQFGYV AMFSVAWPLA ACCFLVNNWV ELRSDALKIA ISSRRPIPWR TDSIGP WLT ALSFLSWLGS ITSSAIVYLC SNSKNGTQGE ASPLKAWGLL LSILFAEHFY LVVQLAVRFV LSKLDSPGLQ KERKERF QT KKRLLQENLG QDAAEEAAAP GIEHSEKITR EALEEEARQA SIRGHGTPEE MFWQRQRGMQ ETIEIGRRMI EQQLAAGK N GKKSAPAVPS EKAS UniProtKB: Plasma membrane channel protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.3 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 5 mM Hepes 7.6, 150 mM NaCl, 0.03% DDM, 2 mM EGTA |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: at 5mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 9 / Number real images: 3351 / Average exposure time: 9.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6qm6: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)