-Search query
-Search result
Showing all 47 items for (author: meyer & eh)

EMDB-15806: 
Cryo-EM structure of the plant 80S ribosome
Method: single particle / : Smirnova J, Loerke J, Kleinau G, Schmidt A, Buerger J, Meyer EH, Mielke T, Scheerer P, Bock R, Spahn CMT, Zoschke R

EMDB-15674: 
Cryo-EM structure of the plant 40S subunit
Method: single particle / : Smirnova J, Loerke J, Kleinau G, Schmidt A, Buerger J, Meyer EH, Mielke T, Scheerer P, Bock R, Spahn CMT, Zoschke R

EMDB-15773: 
Cryo-EM structure of the plant 60S subunit
Method: single particle / : Smirnova J, Loerke J, Kleinau G, Schmidt A, Buerger J, Meyer EH, Mielke T, Scheerer P, Bock R, Spahn CMT, Zoschke R

EMDB-26719: 
FMC63 scFv in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-26720: 
SJ25C1 Fab in complex with soluble CD19
Method: single particle / : Meyerson J, He C

PDB-7urv: 
FMC63 scFv in complex with soluble CD19
Method: single particle / : Meyerson J, He C

PDB-7urx: 
SJ25C1 Fab in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-14964: 
HOPS tethering complex from yeast, composite map
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14965: 
HOPS tethering complex from yeast, consensus map covering the upper part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14966: 
HOPS tethering complex from yeast, consensus map covering the bottom part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14967: 
HOPS tethering complex from yeast, local refinement map of the SNARE-binding module
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14968: 
HOPS tethering complex from yeast, local refinement map of the backbone part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14969: 
HOPS tethering complex from yeast, local refinement map of the bottom part of the complex (Vps18)
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14970: 
HOPS tethering complex from yeast, local refinement map of the bottom part of the complex (Vps39)
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

PDB-7zu0: 
HOPS tethering complex from yeast
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-25076: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

EMDB-25077: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

PDB-7l6o: 
Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23518: 
Cryo-EM map of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lu9: 
Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23519: 
Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lua: 
Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23152: 
Cryo-electron microscopy reconstruction of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer
Method: single particle / : Edwards RJ, Acharya P

EMDB-23153: 
Cryo-electron microscopy local refinement of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer
Method: single particle / : Edwards RJ, Acharya P

EMDB-23149: 
Cryo-electron microscopy reconstruction of antibody DH898.1 Fab-dimer bound near the CD4 binding site of HIV Env SOSIP trimer CH848 10.17
Method: single particle / : Edwards RJ, Acharya P

EMDB-23124: 
Cryo-electron microcospy reconstruction of CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 HIV Env
Method: single particle / : Edwards RJ, Acharya P

EMDB-23145: 
Cryo-electron microscopy reconstruction of locally refined antibody DH898.1 Fab-dimer
Method: single particle / : Edwards RJ, Acharya P

PDB-7l6m: 
Cryo-EM structure of DH898.1 Fab-dimer from local refinement of the Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23094: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-23095: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-23097: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l02: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l06: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l09: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-22221: 
SARS-CoV-2 HexaPro S One RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS

EMDB-22222: 
SARS-CoV-2 HexaPro S Two RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS

PDB-6xkl: 
SARS-CoV-2 HexaPro S One RBD up
Method: single particle / : Wrapp D, Hsieh CL, Goldsmith JA, McLellan JS
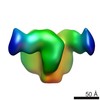
EMDB-8507: 
92BR SOSIP.664 trimer in complex with DH270.1 Fab
Method: single particle / : Fera D, Harrison SC
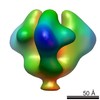
EMDB-5544: 
Molecular structure of the native HIV-1 Env trimer bound to VHH A12: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5551: 
Molecular structure of the native HIV-1 Env trimer bound to VHH A12: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S
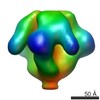
EMDB-5552: 
Molecular structure of the native HIV-1 Env trimer bound to m36: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5553: 
Molecular structure of the native HIV-1 Env trimer bound to m36: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5554: 
Molecular structure of the native HIV-1 Env trimer bound to m36 and sCD4: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5555: 
Molecular structure of the native HIV-1 Env trimer bound to m36 and sCD4: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5423: 
Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-terminal Type IV Pilin Helices
Method: helical / : Yu S, Goforth C, Meyer C, Rachel R, Wirth R, Schroeder G, Egelman EH

EMDB-1615: 
Three-dimensional structure of YidC bound to the translating ribosome
Method: single particle / : Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N

EMDB-1616: 
Three-dimensional structure of Oxa1 bound to the translating ribosome
Method: single particle / : Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
