+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8507 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 92BR SOSIP.664 trimer in complex with DH270.1 Fab | |||||||||
 Map data Map data | DH270.1 Fab in complex with 92BR SOSIP.664 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of establishment of T cell polarity / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / apoptotic process ...: / positive regulation of establishment of T cell polarity / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / apoptotic process / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 29.4 Å | |||||||||
 Authors Authors | Fera D / Harrison SC | |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2017 Journal: Sci Transl Med / Year: 2017Title: Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Authors: Mattia Bonsignori / Edward F Kreider / Daniela Fera / R Ryan Meyerhoff / Todd Bradley / Kevin Wiehe / S Munir Alam / Baptiste Aussedat / William E Walkowicz / Kwan-Ki Hwang / Kevin O ...Authors: Mattia Bonsignori / Edward F Kreider / Daniela Fera / R Ryan Meyerhoff / Todd Bradley / Kevin Wiehe / S Munir Alam / Baptiste Aussedat / William E Walkowicz / Kwan-Ki Hwang / Kevin O Saunders / Ruijun Zhang / Morgan A Gladden / Anthony Monroe / Amit Kumar / Shi-Mao Xia / Melissa Cooper / Mark K Louder / Krisha McKee / Robert T Bailer / Brendan W Pier / Claudia A Jette / Garnett Kelsoe / Wilton B Williams / Lynn Morris / John Kappes / Kshitij Wagh / Gift Kamanga / Myron S Cohen / Peter T Hraber / David C Montefiori / Ashley Trama / Hua-Xin Liao / Thomas B Kepler / M Anthony Moody / Feng Gao / Samuel J Danishefsky / John R Mascola / George M Shaw / Beatrice H Hahn / Stephen C Harrison / Bette T Korber / Barton F Haynes /    Abstract: A preventive HIV-1 vaccine should induce HIV-1-specific broadly neutralizing antibodies (bnAbs). However, bnAbs generally require high levels of somatic hypermutation (SHM) to acquire breadth, and ...A preventive HIV-1 vaccine should induce HIV-1-specific broadly neutralizing antibodies (bnAbs). However, bnAbs generally require high levels of somatic hypermutation (SHM) to acquire breadth, and current vaccine strategies have not been successful in inducing bnAbs. Because bnAbs directed against a glycosylated site adjacent to the third variable loop (V3) of the HIV-1 envelope protein require limited SHM, the V3-glycan epitope is an attractive vaccine target. By studying the cooperation among multiple V3-glycan B cell lineages and their coevolution with autologous virus throughout 5 years of infection, we identify key events in the ontogeny of a V3-glycan bnAb. Two autologous neutralizing antibody lineages selected for virus escape mutations and consequently allowed initiation and affinity maturation of a V3-glycan bnAb lineage. The nucleotide substitution required to initiate the bnAb lineage occurred at a low-probability site for activation-induced cytidine deaminase activity. Cooperation of B cell lineages and an improbable mutation critical for bnAb activity defined the necessary events leading to breadth in this V3-glycan bnAb lineage. These findings may, in part, explain why initiation of V3-glycan bnAbs is rare, and suggest an immunization strategy for inducing similar V3-glycan bnAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
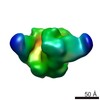
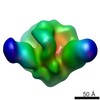
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8507.map.gz emd_8507.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8507-v30.xml emd-8507-v30.xml emd-8507.xml emd-8507.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8507.png emd_8507.png | 15.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8507 http://ftp.pdbj.org/pub/emdb/structures/EMD-8507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8507 | HTTPS FTP |
-Related structure data
| Related structure data |  5tplC  5tppC  5tqaC  5trpC  5u0rC  5u0uC  5u15C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8507.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8507.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DH270.1 Fab in complex with 92BR SOSIP.664 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
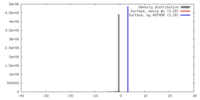
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 92BR HIV Env SOSIP and DH720.1 Fab
| Entire | Name: 92BR HIV Env SOSIP and DH720.1 Fab |
|---|---|
| Components |
|
-Supramolecule #1: 92BR HIV Env SOSIP and DH720.1 Fab
| Supramolecule | Name: 92BR HIV Env SOSIP and DH720.1 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pVRC-8400 Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pVRC-8400 |
-Macromolecule #1: 92BR SOSIP.664
| Macromolecule | Name: 92BR SOSIP.664 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Sequence | String: KDNLWVTVYY GVPVWKEATT TLFCASDAKA YKAEVHNVWA THACVPTDPN PQEIVLENVT ENFNMWKNNM VEQMHEDIIS LWDQSLKPCV KLTPLCVTLN CIDLNNSTNN NNSSGVKTGI DKGEIKNCSF NTTTSVKDKE KKEYALFYNL DVVQIGNDNT SYRLTSCNTS ...String: KDNLWVTVYY GVPVWKEATT TLFCASDAKA YKAEVHNVWA THACVPTDPN PQEIVLENVT ENFNMWKNNM VEQMHEDIIS LWDQSLKPCV KLTPLCVTLN CIDLNNSTNN NNSSGVKTGI DKGEIKNCSF NTTTSVKDKE KKEYALFYNL DVVQIGNDNT SYRLTSCNTS VITQACPKVT FEPIPIHYCT PAGYAILKCN GKKFNGTGPC TNVSTVQCTH GIKPVVSTQL LLNGSLAEED IVIRSENLTN NAKTIIVQLK DPVDINCTRP NNNTRKSIHI GPGRAFYATG DIIGDIRQAH CNLSRAQWND TLSKIVTKLR EQFENKTIKF QPPSGGDPEI VFHSFNCGGE FFYCNTTQLF NSTWTNNTEG TSNTTGNDTI TLPCRIKQIV NMWQEVGKAM YAPPIKGKIK CSSNITGLLL TRDGGNNEMN TTEIFRPGGG DMRDNWRSEL YKYKVVRIEP LGIAPTRCKR RVVGRRRRRR AVGTLGAMFL GFLGAAGSTM GAASVALTVQ ARQLLSGIVQ QQNNLLRAPE AQQHMLQLTV WGIKQLQARV LAVERYLGDQ QLLGIWGCSG KLICCTTVPW NTSWSNKSLD DIWTNMTWME WKREIDNYTS LIYTLIEESQ RQQEKNEQEL LELD |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #2: DH270.1 Fab Heavy Chain
| Macromolecule | Name: DH270.1 Fab Heavy Chain / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE MKKPGASVRV SCKASGYTFT DYYIHWVRQA PGQGPEWMGW INPSTGRTNS PQKFQGRVTM TRDTSISTAY MDLNRLTSDD TAMYYCTTGG WIGLYSDTSG YPNFDYWGQG TLVTVSGAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS ...String: QVQLVQSGAE MKKPGASVRV SCKASGYTFT DYYIHWVRQA PGQGPEWMGW INPSTGRTNS PQKFQGRVTM TRDTSISTAY MDLNRLTSDD TAMYYCTTGG WIGLYSDTSG YPNFDYWGQG TLVTVSGAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DKHHHHHH |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #3: DH270.1 Fab Light Chain
| Macromolecule | Name: DH270.1 Fab Light Chain / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTNYDVG SYNLVSWYQQ HPGKVPKYII YEVNKRPSGV SNRFSGSKSG NTASLTISGL QAEDEATYYC CSYAGSSIIF FGGGTKLTVI GQPKGAPSVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK ...String: QSALTQPASV SGSPGQSITI SCTGTNYDVG SYNLVSWYQQ HPGKVPKYII YEVNKRPSGV SNRFSGSKSG NTASLTISGL QAEDEATYYC CSYAGSSIIF FGGGTKLTVI GQPKGAPSVT LFPPSSEELQ ANKATLVCLI SDFYPGAVTV AWKADSSPVK AGVETTTPSK QSNNKYAASS YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM HEPES, pH 8.0, 150 mM NaCl, 0.02% sodium azide |
| Staining | Type: NEGATIVE / Material: 2% uranyl formate |
| Grid | Model: Electron Microscopy Sciences / Material: COPPER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 29.4 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 5419 |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)