+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23518 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of DH851.3 bound to HIV-1 CH505 Env | |||||||||
 Map data Map data | CryoEM map of DH851.3 bound to HIV-1 CH505 Env | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fab-dimerized / glycan-reactive / antibodies / HIV-1 / DH898.1 / Fab-dimer / glycan-reactive neutralizing antibody / macaque HIV-1 vaccine-induced / B cell lineage / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Edwards RJ / Manne K / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler ...Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler Evangelous / Bhavna Hora / Madison Berry / A Yousef Abuahmad / Jordan Sprenz / Margaret Deyton / Victoria Stalls / Megan Kopp / Allen L Hsu / Mario J Borgnia / Guillaume B E Stewart-Jones / Matthew S Lee / Naomi Bronkema / M Anthony Moody / Kevin Wiehe / Todd Bradley / S Munir Alam / Robert J Parks / Andrew Foulger / Thomas Oguin / Gregory D Sempowski / Mattia Bonsignori / Celia C LaBranche / David C Montefiori / Michael Seaman / Sampa Santra / John Perfect / Joseph R Francica / Geoffrey M Lynn / Baptiste Aussedat / William E Walkowicz / Richard Laga / Garnett Kelsoe / Kevin O Saunders / Daniela Fera / Peter D Kwong / Robert A Seder / Alberto Bartesaghi / George M Shaw / Priyamvada Acharya / Barton F Haynes /   Abstract: Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) ...Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) as a model antigen. 2G12 is a broadly neutralizing Ab (bnAb) that targets a conserved glycan patch on Env of geographically diverse HIV-1 strains using a unique heavy-chain (V) domain-swapped architecture that results in fragment antigen-binding (Fab) dimerization. Here, we describe HIV-1 Env Fab-dimerized glycan (FDG)-reactive bnAbs without V-swapped domains from simian-human immunodeficiency virus (SHIV)-infected macaques. FDG Abs also recognized cell-surface glycans on diverse pathogens, including yeast and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike. FDG precursors were expanded by glycan-bearing immunogens in macaques and were abundant in HIV-1-naive humans. Moreover, FDG precursors were predominately mutated IgMIgDCD27, thus suggesting that they originated from a pool of antigen-experienced IgM or marginal zone B cells. #1:  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Fab-dimerized glycan-reactive antibodies neutralize HIV and are prevalent in humans and rhesus macaques Authors: Acharya P / Williams W / Henderson R / Janowska K / Manne K / Parks R / Deyton M / Sprenz J / Stalls V / Kopp M / Mansouri K / Edwards RJ / Meyerhoff RR / Oguin T / Sempowski G / Saunders K / Haynes BF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23518.map.gz emd_23518.map.gz | 25.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23518-v30.xml emd-23518-v30.xml emd-23518.xml emd-23518.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23518.png emd_23518.png | 70.9 KB | ||
| Filedesc metadata |  emd-23518.cif.gz emd-23518.cif.gz | 7.7 KB | ||
| Others |  emd_23518_half_map_1.map.gz emd_23518_half_map_1.map.gz emd_23518_half_map_2.map.gz emd_23518_half_map_2.map.gz | 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23518 http://ftp.pdbj.org/pub/emdb/structures/EMD-23518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23518 | HTTPS FTP |
-Related structure data
| Related structure data |  7lu9MC  6vtuC  6xrjC  7l02C  7l06C  7l09C  7l6mC  7l6oC  7luaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23518.map.gz / Format: CCP4 / Size: 27.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23518.map.gz / Format: CCP4 / Size: 27.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of DH851.3 bound to HIV-1 CH505 Env | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0665 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
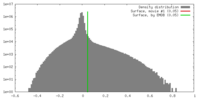
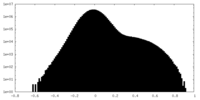
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: CryoEM half-map B of DH851.3 bound to HIV-1 CH505 Env
| File | emd_23518_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half-map B of DH851.3 bound to HIV-1 CH505 Env | ||||||||||||
| Projections & Slices |
| ||||||||||||
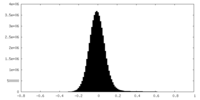
| Density Histograms |
-Half map: CryoEM half-map A of DH851.3 bound to HIV-1 CH505 Env
| File | emd_23518_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half-map A of DH851.3 bound to HIV-1 CH505 Env | ||||||||||||
| Projections & Slices |
| ||||||||||||
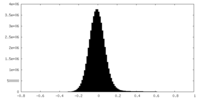
| Density Histograms |
- Sample components
Sample components
-Entire : DH851.3 bound to HIV-1 CH505 Env
| Entire | Name: DH851.3 bound to HIV-1 CH505 Env |
|---|---|
| Components |
|
-Supramolecule #1: DH851.3 bound to HIV-1 CH505 Env
| Supramolecule | Name: DH851.3 bound to HIV-1 CH505 Env / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: DH851.3 light chain
| Macromolecule | Name: DH851.3 light chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.127463 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALTQPPSVSK SLEQSVTISC TGTTTGNSVS WYQCHSGTAP RLLIYDVNKR PSGVSDRFSG SKSHNTASLT IFGLQAEDEA DYYCGSYGS GGSLLFGGGT RLTVLGQP(UNK)K ASPTVTLFPP SSEELQANKA TLVCLISDFY PGVVKVAWKA DGSAVNA GV ETTTPSKQSN ...String: ALTQPPSVSK SLEQSVTISC TGTTTGNSVS WYQCHSGTAP RLLIYDVNKR PSGVSDRFSG SKSHNTASLT IFGLQAEDEA DYYCGSYGS GGSLLFGGGT RLTVLGQP(UNK)K ASPTVTLFPP SSEELQANKA TLVCLISDFY PGVVKVAWKA DGSAVNA GV ETTTPSKQSN NKYAASSYLS LTSDQWKSHK SYSCQVTHEG STVEKTVAPA EC |
-Macromolecule #2: DH851.3 heavy chain
| Macromolecule | Name: DH851.3 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.451617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVTLMESGPA LVKVTQTLAV TCTFSGFSIR DSGKGVAWIR QPPGGALEWL TSIYWDDTKY HDTSLKPRLT IFRDTSQTQV ILILTNMAP LDTATYYCGR INNGGGWKDH IDFWGPGLLV TVSSASTKGP SVFPLAPSSR STSESTAALG CLVKDYFPEP V TVSWNSGS ...String: QVTLMESGPA LVKVTQTLAV TCTFSGFSIR DSGKGVAWIR QPPGGALEWL TSIYWDDTKY HDTSLKPRLT IFRDTSQTQV ILILTNMAP LDTATYYCGR INNGGGWKDH IDFWGPGLLV TVSSASTKGP SVFPLAPSSR STSESTAALG CLVKDYFPEP V TVSWNSGS LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYVCNV NHKPSNTKVD KRVEIKTCGG |
-Macromolecule #3: DH851.3 heavy chain
| Macromolecule | Name: DH851.3 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.89093 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVTLMESGPA LVKVTQTLAV TCTFSGFSIR DSGKGVAWIR QPPGGALEWL TSIYWDDTKY HDTSLKPRLT IFRDTSQTQV ILILTNMAP LDTATYYCGR INNGGGWKDH IDFWGPGLLV TVSSASTKGP SVFPLAPSSR STSESTAALG CLVKDYFPEP V TVSWNSGS ...String: QVTLMESGPA LVKVTQTLAV TCTFSGFSIR DSGKGVAWIR QPPGGALEWL TSIYWDDTKY HDTSLKPRLT IFRDTSQTQV ILILTNMAP LDTATYYCGR INNGGGWKDH IDFWGPGLLV TVSSASTKGP SVFPLAPSSR STSESTAALG CLVKDYFPEP V TVSWNSGS LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYVCNV NHKPSNTKVD KRVE |
-Macromolecule #4: DH851.3 light chain
| Macromolecule | Name: DH851.3 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.824127 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALTQPPSVSK SLEQSVTISC TGTTTGNSVS WYQCHSGTAP RLLIYDVNKR PSGVSDRFSG SKSHNTASLT IFGLQAEDEA DYYCGSYGS GGSLLFGGGT RLTVLGQP(UNK)K ASPTVTLFPP SSEELQANKA TLVCLISDFY PGVVKVAWKA DGSAVNA GV ETTTPSKQSN ...String: ALTQPPSVSK SLEQSVTISC TGTTTGNSVS WYQCHSGTAP RLLIYDVNKR PSGVSDRFSG SKSHNTASLT IFGLQAEDEA DYYCGSYGS GGSLLFGGGT RLTVLGQP(UNK)K ASPTVTLFPP SSEELQANKA TLVCLISDFY PGVVKVAWKA DGSAVNA GV ETTTPSKQSN NKYAASSYLS LTSDQWKSHK SYSCQVTHEG STVEKTVAP |
-Macromolecule #5: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.134512 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLKAIEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #6: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 51.898062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ENLWVTVYYG VPVWKEAKTT LFCASDAKAY EKEVHNVWAT HACVPTDPNP QEMVLKNVTE NFNMWKNDMV DQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS V ITQACPKV ...String: ENLWVTVYYG VPVWKEAKTT LFCASDAKAY EKEVHNVWAT HACVPTDPNP QEMVLKNVTE NFNMWKNDMV DQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS V ITQACPKV SFDPIPIHYC APAGYAILKC NNKTFTGTGP CNNVSTVQCT HGIKPVVSTQ LLLNGSLAEG EIIIRSENIT NN VKTIIVH LNESVKIECT RPNNKTRTSI RIGPGQWFYA TGQVIGDIRE AYCNINESKW NETLQRVSKK LKEYFPHKNI TFQ PSSGGD LEITTHSFNC GGEFFYCNTS SLFNRTYMAN STDMANSTET NSTRTITIHC RIKQIINMWQ EVGRAMYAPP IAGN ITCIS NITGLLLTRD GGKNNTETFR PGGGNMKDNW RSELYKYKVV KIEPLGVAPT RCKRRV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 12 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: LEICA EM GP / Details: blot for 2.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.15 K / Max: 93.15 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 6058 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 53398 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7lu9: |
 Movie
Movie Controller
Controller





















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)