[English] 日本語
 Yorodumi
Yorodumi- EMDB-23519: Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23519 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | |||||||||
 Map data Map data | Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fab-dimerized / glycan-reactive / antibodies / HIV-1 / DH898.1 / Fab-dimer / glycan-reactive neutralizing antibody / macaque HIV-1 vaccine-induced / B cell lineage / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope ...membrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Simian-Human immunodeficiency virus / Simian-Human immunodeficiency virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Edwards RJ / Manne K / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler ...Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler Evangelous / Bhavna Hora / Madison Berry / A Yousef Abuahmad / Jordan Sprenz / Margaret Deyton / Victoria Stalls / Megan Kopp / Allen L Hsu / Mario J Borgnia / Guillaume B E Stewart-Jones / Matthew S Lee / Naomi Bronkema / M Anthony Moody / Kevin Wiehe / Todd Bradley / S Munir Alam / Robert J Parks / Andrew Foulger / Thomas Oguin / Gregory D Sempowski / Mattia Bonsignori / Celia C LaBranche / David C Montefiori / Michael Seaman / Sampa Santra / John Perfect / Joseph R Francica / Geoffrey M Lynn / Baptiste Aussedat / William E Walkowicz / Richard Laga / Garnett Kelsoe / Kevin O Saunders / Daniela Fera / Peter D Kwong / Robert A Seder / Alberto Bartesaghi / George M Shaw / Priyamvada Acharya / Barton F Haynes /   Abstract: Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) ...Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) as a model antigen. 2G12 is a broadly neutralizing Ab (bnAb) that targets a conserved glycan patch on Env of geographically diverse HIV-1 strains using a unique heavy-chain (V) domain-swapped architecture that results in fragment antigen-binding (Fab) dimerization. Here, we describe HIV-1 Env Fab-dimerized glycan (FDG)-reactive bnAbs without V-swapped domains from simian-human immunodeficiency virus (SHIV)-infected macaques. FDG Abs also recognized cell-surface glycans on diverse pathogens, including yeast and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike. FDG precursors were expanded by glycan-bearing immunogens in macaques and were abundant in HIV-1-naive humans. Moreover, FDG precursors were predominately mutated IgMIgDCD27, thus suggesting that they originated from a pool of antigen-experienced IgM or marginal zone B cells. #1:  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Fab-dimerized glycan-reactive antibodies neutralize HIV and are prevalent in humans and rhesus macaques Authors: Acharya P / Williams W / Henderson R / Janowska K / Manne K / Parks R / Deyton M / Sprenz J / Stalls V / Kopp M / Mansouri K / Edwards RJ / Meyerhoff RR / Oguin T / Sempowski G / Saunders K / Haynes BF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23519.map.gz emd_23519.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23519-v30.xml emd-23519-v30.xml emd-23519.xml emd-23519.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23519.png emd_23519.png | 129.5 KB | ||
| Filedesc metadata |  emd-23519.cif.gz emd-23519.cif.gz | 7.6 KB | ||
| Others |  emd_23519_half_map_1.map.gz emd_23519_half_map_1.map.gz emd_23519_half_map_2.map.gz emd_23519_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23519 http://ftp.pdbj.org/pub/emdb/structures/EMD-23519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23519 | HTTPS FTP |
-Related structure data
| Related structure data |  7luaMC  6vtuC  6xrjC  7l02C  7l06C  7l09C  7l6mC  7l6oC  7lu9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23519.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23519.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
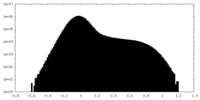
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0665 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
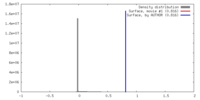
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Cryo-EM map of DH898.1 Fab-dimer bound near the...
| File | emd_23519_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
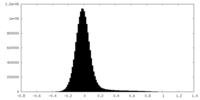
| Density Histograms |
-Half map: Cryo-EM half-map B of DH898.1 Fab-dimer bound near...
| File | emd_23519_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half-map B of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
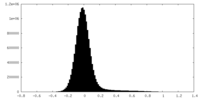
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding...
| Entire | Name: Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer. |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding...
| Supramolecule | Name: Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: CH848 SOSIP gp120
| Macromolecule | Name: CH848 SOSIP gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 52.06193 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKEAKT TLFCASDARA YEKEVHNVWA THACVPTDPS PQELVLGNVT ENFNMWKNDM VDQMHEDIIS LWDQSLKPC VKLTPLCVTL ICSNATVKNG TVEEMKNCSF NTTTEIRDKE KKEYALFYKP DIVPLSETNN TSEYRLINCN T SACTQACP ...String: AENLWVTVYY GVPVWKEAKT TLFCASDARA YEKEVHNVWA THACVPTDPS PQELVLGNVT ENFNMWKNDM VDQMHEDIIS LWDQSLKPC VKLTPLCVTL ICSNATVKNG TVEEMKNCSF NTTTEIRDKE KKEYALFYKP DIVPLSETNN TSEYRLINCN T SACTQACP KVTFEPIPIH YCAPAGYAIL KCNDETFNGT GPCSNVSTVQ CTHGIRPVVS TQLLLNGSLA EKEIVIRSEN LT NNAKIII VHLHTPVEIV CTRPNNNTRK SVRIGPGQTF YATGDIIGDI KQAHCNISEE KWNDTLQKVG IELQKHFPNK TIK YNQSAG GDMEITTHSF NCGGEFFYCN TSNLFNGTYN GTYISTNSSA NSTSTITLQC RIKQIINMWQ GVGRCMYAPP IAGN ITCRS NITGLLLTRD GGTNSNETET FRPAGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Env polyprotein
| Macromolecule | Name: Env polyprotein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian-Human immunodeficiency virus Simian-Human immunodeficiency virus |
| Molecular weight | Theoretical: 16.578803 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FLGFLGAAGS TMGAASMTLT VQARNLLSGI VQQQSNLLRA PEAQQHLLKL TVWGIKQLQA RVLAVERYLR DQQLLGIWGC SGKLICCTN VPWNSSWSNR NLSEIWDNMT WLQWDKEISN YTQIIYGLLE ESQNQQEKNE QDLLALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: DH898.1 light chain
| Macromolecule | Name: DH898.1 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.55325 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQTPLS LSVTPGEPAS LSCRSSASLL HGNGNTYLHW YLRKAGQSPQ LLIFGGSKRV PGISDRFIGS GAGTNFTLKI SSVEADDVG FYYCAQGVAF PWTFGQGTKV EIKRAVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGVL K TGNSQESV ...String: EIVMTQTPLS LSVTPGEPAS LSCRSSASLL HGNGNTYLHW YLRKAGQSPQ LLIFGGSKRV PGISDRFIGS GAGTNFTLKI SSVEADDVG FYYCAQGVAF PWTFGQGTKV EIKRAVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGVL K TGNSQESV TEQDSKDNTY SLSSTLTLSN TDYQSHNVYA CEVTHQGLSS PVTKSFNRGE |
-Macromolecule #4: DH898.1 heavy chain
| Macromolecule | Name: DH898.1 heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.596594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QDLLLQSGAE VREPGASVTV SCQASNYTFP DYYIHWVRLV PGQGLEWLGE MKPKVGVTNV SKKIRDRLFM TADTSTDTAY MVLSALTPG DTAIYYCTRL EPDFLSGWAH WGKGVLVTVS PASTKGPSVF PLAPSSRSTS ESTAALGCLV KDYFPEPVTV S WNSGSLTS ...String: QDLLLQSGAE VREPGASVTV SCQASNYTFP DYYIHWVRLV PGQGLEWLGE MKPKVGVTNV SKKIRDRLFM TADTSTDTAY MVLSALTPG DTAIYYCTRL EPDFLSGWAH WGKGVLVTVS PASTKGPSVF PLAPSSRSTS ESTAALGCLV KDYFPEPVTV S WNSGSLTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYVCNVNHK PSNTKVDKRV E |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 17 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: LEICA EM GP / Details: blot for 2.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.15 K / Max: 93.15 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3230 / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7lua: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)