[English] 日本語
 Yorodumi
Yorodumi- EMDB-2361: Electron cryo-EM of the full-length Thermus thermophilus DNA gyra... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2361 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-EM of the full-length Thermus thermophilus DNA gyrase in complex with a 155bp DNA and ciprofloxacin | |||||||||
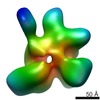
 Map data Map data | Recontruction of the full length Thermus thermophilus DNA gyrase bound to DNA. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA gyrase / DNA topoisomerase / DNA supercoiling | |||||||||
| Biological species |   Thermus thermophilus (bacteria) / Thermus thermophilus (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Papillon J / Menetret JF / Batisse C / Helye R / Schultz P / Potier N / Lamour V | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2013 Journal: Nucleic Acids Res / Year: 2013Title: Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type 2A DNA topoisomerase. Authors: Julie Papillon / Jean-François Ménétret / Claire Batisse / Reynald Hélye / Patrick Schultz / Noëlle Potier / Valérie Lamour /  Abstract: Type 2A DNA topoisomerases (Topo2A) remodel DNA topology during replication, transcription and chromosome segregation. These multisubunit enzymes catalyze the transport of a double-stranded DNA ...Type 2A DNA topoisomerases (Topo2A) remodel DNA topology during replication, transcription and chromosome segregation. These multisubunit enzymes catalyze the transport of a double-stranded DNA through a transient break formed in another duplex. The bacterial DNA gyrase, a target for broad-spectrum antibiotics, is the sole Topo2A enzyme able to introduce negative supercoils. We reveal here for the first time the architecture of the full-length Thermus thermophilus DNA gyrase alone and in a cleavage complex with a 155 bp DNA duplex in the presence of the antibiotic ciprofloxacin, using cryo-electron microscopy. The structural organization of the subunits of the full-length DNA gyrase points to a central role of the ATPase domain acting like a 'crossover trap' that may help to sequester the DNA positive crossover before strand passage. Our structural data unveil how DNA is asymmetrically wrapped around the gyrase-specific C-terminal β-pinwheel domains and guided to introduce negative supercoils through cooperativity between the ATPase and β-pinwheel domains. The overall conformation of the drug-induced DNA binding-cleavage complex also suggests that ciprofloxacin traps a DNA pre-transport conformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2361.map.gz emd_2361.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2361-v30.xml emd-2361-v30.xml emd-2361.xml emd-2361.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2361.png emd_2361.png | 48.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2361 http://ftp.pdbj.org/pub/emdb/structures/EMD-2361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2361 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2361.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2361.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Recontruction of the full length Thermus thermophilus DNA gyrase bound to DNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.92 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA-bound complex of Thermus thermophilus DNA gyrase with a 155bp...
| Entire | Name: DNA-bound complex of Thermus thermophilus DNA gyrase with a 155bp DNA in presence of ADPNP and ciprofoxacin. |
|---|---|
| Components |
|
-Supramolecule #1000: DNA-bound complex of Thermus thermophilus DNA gyrase with a 155bp...
| Supramolecule | Name: DNA-bound complex of Thermus thermophilus DNA gyrase with a 155bp DNA in presence of ADPNP and ciprofoxacin. type: sample / ID: 1000 / Details: monodisperse complex / Oligomeric state: dimer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 413 KDa / Theoretical: 413 KDa / Method: native mass spectrometry |
-Macromolecule #1: DNA gyrase
| Macromolecule | Name: DNA gyrase / type: protein_or_peptide / ID: 1 / Name.synonym: bacterial DNA topoisomerase 2A Details: The two subinits of the DNA gyrase were fused for structural stability. Oligomeric state: dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 321 KDa / Theoretical: 321 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: linear DNA
| Macromolecule | Name: linear DNA / type: dna / ID: 2 Details: A 155bp DNA was used to form the DNA-bound complex. ADPNP (a non hydrolyzable analog of ATP) and ciprofloxacin (quinolone antibiotic) were added to stabilize the complex Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.150 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20mM Hepes, 100 mM NaCl, 5mM MgCl2, 1mM DTT |
| Grid | Details: Quantifoil R 2/2 holey carbon copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Method: Plunging immediately after blotting |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Date | Dec 11, 2011 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Number real images: 900 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 59000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: -1.0 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase flipping (each particle) |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2 / Number images used: 41500 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















