[English] 日本語
 Yorodumi
Yorodumi- EMDB-1615: Three-dimensional structure of YidC bound to the translating ribosome -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional structure of YidC bound to the translating ribosome | |||||||||
 Map data Map data | This map shows YidC bound to the tunnel exit region of an ribosome nascent chain complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein insertion / ribosome / translation | |||||||||
| Function / homology | : / protein insertion into membrane Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.4 Å | |||||||||
 Authors Authors | Kohler R / Boehringer D / Greber B / Bingel-Erlenmeyer R / Collinson I / Schaffitzel C / Ban N | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2009 Journal: Mol Cell / Year: 2009Title: YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Authors: Rebecca Kohler / Daniel Boehringer / Basil Greber / Rouven Bingel-Erlenmeyer / Ian Collinson / Christiane Schaffitzel / Nenad Ban /  Abstract: The YidC/Oxa1/Alb3 family of membrane proteins facilitates the insertion and assembly of membrane proteins in bacteria, mitochondria, and chloroplasts. Here we present the structures of both ...The YidC/Oxa1/Alb3 family of membrane proteins facilitates the insertion and assembly of membrane proteins in bacteria, mitochondria, and chloroplasts. Here we present the structures of both Escherichia coli YidC and Saccharomyces cerevisiae Oxa1 bound to E. coli ribosome nascent chain complexes determined by cryo-electron microscopy. Dimers of YidC and Oxa1 are localized above the exit of the ribosomal tunnel. Crosslinking experiments show that the ribosome specifically stabilizes the dimeric state. Functionally important and conserved transmembrane helices of YidC and Oxa1 were localized at the dimer interface by cysteine crosslinking. Both Oxa1 and YidC dimers contact the ribosome at ribosomal protein L23 and conserved rRNA helices 59 and 24, similarly to what was observed for the nonhomologous SecYEG translocon. We suggest that dimers of the YidC and Oxa1 proteins form insertion pores and share a common overall architecture with the SecY monomer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1615.map.gz emd_1615.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1615-v30.xml emd-1615-v30.xml emd-1615.xml emd-1615.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1615_1615.png 1615_1615.png | 175.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1615 http://ftp.pdbj.org/pub/emdb/structures/EMD-1615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1615 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1615.map.gz / Format: CCP4 / Size: 4.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1615.map.gz / Format: CCP4 / Size: 4.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map shows YidC bound to the tunnel exit region of an ribosome nascent chain complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.76 Å | ||||||||||||||||||||||||||||||||||||
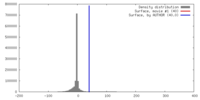
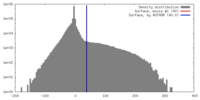
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : YidC bound to a translating ribosome
| Entire | Name: YidC bound to a translating ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: YidC bound to a translating ribosome
| Supramolecule | Name: YidC bound to a translating ribosome / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2.6 MDa / Theoretical: 2.6 MDa |
-Supramolecule #1: Ribosome nascent chain complex
| Supramolecule | Name: Ribosome nascent chain complex / type: complex / ID: 1 / Name.synonym: Translating ribosome / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 2.6 MDa / Theoretical: 2.6 MDa |
-Macromolecule #1: YidC
| Macromolecule | Name: YidC / type: protein_or_peptide / ID: 1 / Name.synonym: Membrane protein insertase / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 62 KDa / Theoretical: 62 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: protein insertion into membrane / InterPro: INTERPRO: IPR013308 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: 200 mesh copper |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each image |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.4 Å / Resolution method: OTHER / Software - Name: Imagic-5 Spider / Number images used: 24395 |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)