+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4319 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | uL23 beta hairpin loop deletion of E.coli ribosome | |||||||||
 Map data Map data | Sharpened map of E.coli ribosome with the beta-hairpin of uL23 deleted. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | uL23 / loop deletion / ribosomal tunnel / RIBOSOMAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal large subunit assembly / cytosolic large ribosomal subunit / cytoplasmic translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
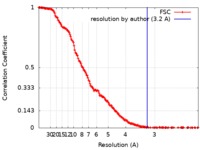
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Kudva R / von Heijne G | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: The shape of the bacterial ribosome exit tunnel affects cotranslational protein folding. Authors: Renuka Kudva / Pengfei Tian / Fátima Pardo-Avila / Marta Carroni / Robert B Best / Harris D Bernstein / Gunnar von Heijne /   Abstract: The ribosome exit tunnel can accommodate small folded proteins, while larger ones fold outside. It remains unclear, however, to what extent the geometry of the tunnel influences protein folding. ...The ribosome exit tunnel can accommodate small folded proteins, while larger ones fold outside. It remains unclear, however, to what extent the geometry of the tunnel influences protein folding. Here, using ribosomes with deletions in loops in proteins uL23 and uL24 that protrude into the tunnel, we investigate how tunnel geometry determines where proteins of different sizes fold. We find that a 29-residue zinc-finger domain normally folding close to the uL23 loop folds deeper in the tunnel in uL23 Δloop ribosomes, while two ~ 100 residue proteins normally folding close to the uL24 loop near the tunnel exit port fold at deeper locations in uL24 Δloop ribosomes, in good agreement with results obtained by coarse-grained molecular dynamics simulations. This supports the idea that cotranslational folding commences once a protein domain reaches a location in the exit tunnel where there is sufficient space to house the folded structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4319.map.gz emd_4319.map.gz | 347.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4319-v30.xml emd-4319-v30.xml emd-4319.xml emd-4319.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4319_fsc.xml emd_4319_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4319.png emd_4319.png | 172.3 KB | ||
| Masks |  emd_4319_msk_1.map emd_4319_msk_1.map | 371.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4319.cif.gz emd-4319.cif.gz | 5.6 KB | ||
| Others |  emd_4319_half_map_1.map.gz emd_4319_half_map_1.map.gz emd_4319_half_map_2.map.gz emd_4319_half_map_2.map.gz | 344.3 MB 344.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4319 http://ftp.pdbj.org/pub/emdb/structures/EMD-4319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4319 | HTTPS FTP |
-Related structure data
| Related structure data |  6fu8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4319.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4319.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of E.coli ribosome with the beta-hairpin of uL23 deleted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4319_msk_1.map emd_4319_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of E.coli ribosome with the beta-hairpin of uL23 deleted.
| File | emd_4319_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of E.coli ribosome with the beta-hairpin of uL23 deleted. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of E.coli ribosome with the beta-hairpin of uL23 deleted.
| File | emd_4319_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of E.coli ribosome with the beta-hairpin of uL23 deleted. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E.coli ribosome with beta-hairpin of uL23 deleted
| Entire | Name: E.coli ribosome with beta-hairpin of uL23 deleted |
|---|---|
| Components |
|
-Supramolecule #1: E.coli ribosome with beta-hairpin of uL23 deleted
| Supramolecule | Name: E.coli ribosome with beta-hairpin of uL23 deleted / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Deposited PDB is of the uL23 with the beta-hairpin loop deleted. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.6 MDa |
-Macromolecule #1: 50S ribosomal protein L23
| Macromolecule | Name: 50S ribosomal protein L23 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.324003 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIREERLLKV LRAPHVSEKA STAMEKSNTI VLKVAKDATK AEIKAAVQKL FEVEVEVVNT LVVKRRSDWK KAYVTLKEGQ NL UniProtKB: Large ribosomal subunit protein uL23 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Support film - Film thickness: 0.02 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.0003 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 3500 / Average electron dose: 1.17 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
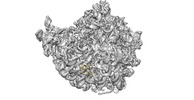
| Output model |  PDB-6fu8: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)