6OE9
 
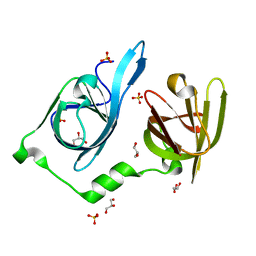 | | Crystal structure of p204 HIN1 domain | | Descriptor: | GLYCEROL, Interferon-activable protein 204, SULFATE ION | | Authors: | Tian, Y, Yin, Q. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of the HIN1 domain of interferon-inducible protein 204.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3O6E
 
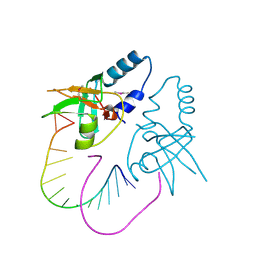 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O3I
 
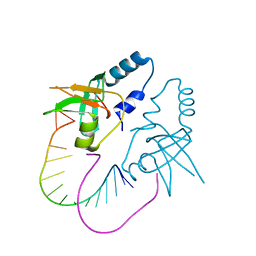 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OH at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*U)-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O7X
 
 | |
3RIU
 
 | |
3O7V
 
 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
4OJV
 
 | | Crystal structure of unliganded yeast PDE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, SULFATE ION, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
7SA4
 
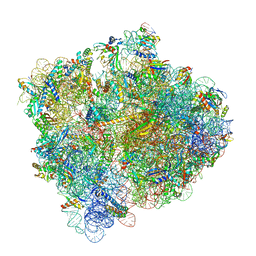 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tian, Y, Zeng, F, Raybarman, A, Carruthers, A, Li, Q, Fatma, S, Huang, R.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Sequential rescue and repair of stalled and damaged ribosome by bacterial PrfH and RtcB.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ESH
 
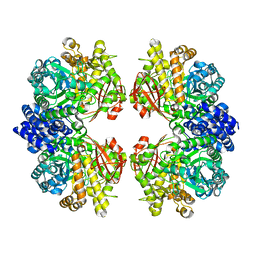 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
7TWD
 
 | | Structure of AAGAB C-terminal dimerization domain | | Descriptor: | Alpha- and gamma-adaptin-binding protein p34, PHOSPHATE ION | | Authors: | Tian, Y, Yin, Q. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Oligomer-to-monomer transition underlies the chaperone function of AAGAB in AP1/AP2 assembly.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4NGD
 
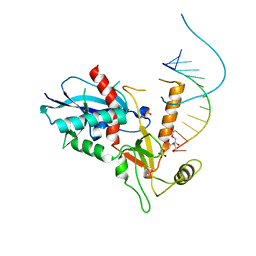 | | Structure of human Dicer Platform-PAZ-Connector Helix cassette in complex with 12-mer siRNA having 5'-p and UU-3' ends (1.95 Angstrom resolution) | | Descriptor: | 5'-R(P*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*UP*U)-3', DI(HYDROXYETHYL)ETHER, Endoribonuclease Dicer, ... | | Authors: | Tian, Y, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2013-11-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Mol.Cell, 53, 2014
|
|
4NH5
 
 | |
2MOZ
 
 | | Structure of the Membrane Protein MerF, a Bacterial Mercury Transporter, Improved by the Inclusion of Chemical Shift Anisotropy Constraints | | Descriptor: | MerF | | Authors: | Tian, Y, Lu, G.J, Marassi, F.M, Opella, S.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
J.Biomol.Nmr, 60, 2014
|
|
2H3H
 
 | |
4NH3
 
 | |
4NHA
 
 | | Structure of human Dicer Platform-PAZ-Connector Helix cassette in complex with 16-mer siRNA having 5'-p and UU-3' ends (3.4 Angstrom resolution) | | Descriptor: | 5'-R(P*GP*CP*GP*UP*UP*GP*GP*CP*CP*AP*AP*CP*GP*CP*UP*U)-3', Endoribonuclease Dicer | | Authors: | Simanshu, D.K, Tian, Y, Ma, J.-B, Patel, D.J. | | Deposit date: | 2013-11-04 | | Release date: | 2014-03-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Mol.Cell, 53, 2014
|
|
4NGG
 
 | |
4NGC
 
 | |
4NH6
 
 | |
4NGF
 
 | |
4NGB
 
 | |
2LK9
 
 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
6VKJ
 
 | |
4WQN
 
 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
