5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
1T0P
 
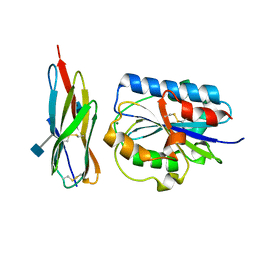 | | Structural Basis of ICAM recognition by integrin alpahLbeta2 revealed in the complex structure of binding domains of ICAM-3 and alphaLbeta2 at 1.65 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, Intercellular adhesion molecule-3, ... | | Authors: | Song, G, Yang, Y.T, Liu, J.H, Shimaoko, M, Springer, T.A, Wang, J.H. | | Deposit date: | 2004-04-12 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3ECR
 
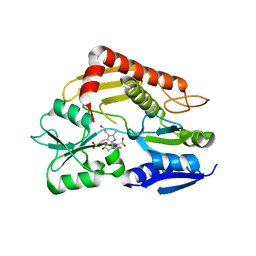 | | Structure of human porphobilinogen deaminase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Song, G, Li, Y, Cheng, C, Zhao, Y, Gao, A, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into acute intermittent porphyria.
Faseb J., 23, 2009
|
|
4M7R
 
 | |
4OKU
 
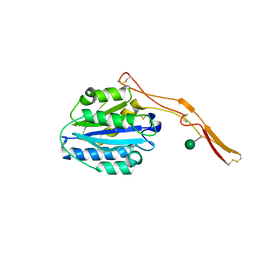 | | Structure of Toxoplasma gondii proMIC2 | | Descriptor: | Micronemal protein MIC2, alpha-D-mannopyranose | | Authors: | Song, G, Springer, T.A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the Toxoplasma gliding motility adhesin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OKR
 
 | | Structures of Toxoplasma gondii MIC2 | | Descriptor: | Micronemal protein MIC2, alpha-D-mannopyranose | | Authors: | Song, G, Springer, T.A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structures of the Toxoplasma gliding motility adhesin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HQO
 
 | | Crystal structure of Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQL
 
 | | Crystal structure of magnesium-loaded Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sporozoite surface protein 2, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQK
 
 | | Crystal structure of Plasmodium falciparum TRAP, P4212 form | | Descriptor: | SULFATE ION, Thrombospondin-related anonymous protein, TRAP | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQN
 
 | | Crystal structure of manganese-loaded Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Sporozoite surface protein 2, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQF
 
 | | Crystal structure of Plasmodium falciparum TRAP, I4 form | | Descriptor: | CHLORIDE ION, Thrombospondin-related anonymous protein, TRAP | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7YQE
 
 | |
6KK7
 
 | | Structure of thermal-stabilised(M6) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
6KK1
 
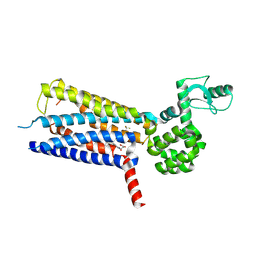 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
6KJV
 
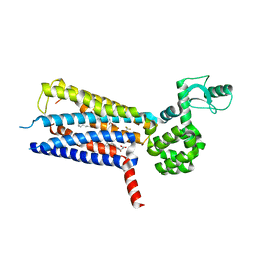 | | Structure of thermal-stabilised(M9) human GLP-1 receptor transmembrane domain | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | Authors: | Yan, X, Li, X, Song, G. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
1ABT
 
 | |
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
3HY4
 
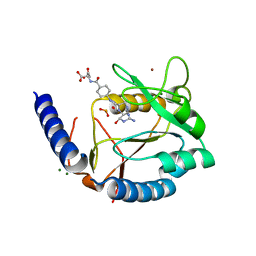 | | Structure of human MTHFS with N5-iminium phosphate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({trans-4-[({(2R,4R,4aS,6S,8aS)-2-amino-4-hydroxy-5-[(phosphonooxy)methyl]decahydropteridin-6-yl}methyl)amino]cyclohexyl}carbonyl)-L-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY6
 
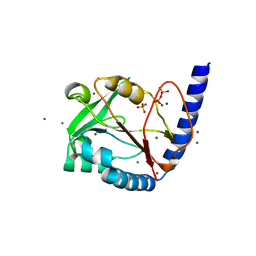 | | Structure of human MTHFS with ADP | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY3
 
 | | Structure of human MTHFS with 10-formyltetrahydrofolate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({4-[{[(2R,4S,4aR,6S,8aS)-2-amino-4-hydroxydecahydropteridin-6-yl]methyl}(formyl)amino]phenyl}carbonyl)-D-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
