1MPL
 
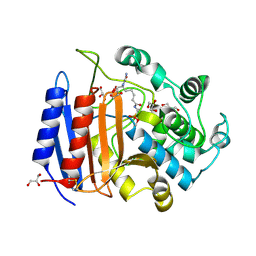 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
1PW8
 
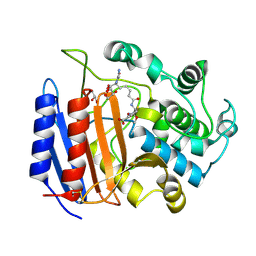 | | Covalent Acyl Enzyme Complex Of The R61 DD-Peptidase with A Highly Specific Cephalosporin | | Descriptor: | (6R,7R)-3-[(ACETYLOXY)METHYL]-7-{[(6S)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-8-OXO-5-THIA-1-AZABICYCLO[4.2.0]OCTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1SDE
 
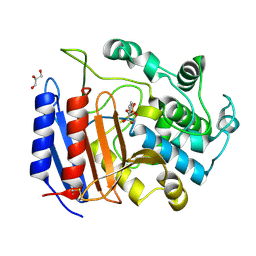 | | Toward Better Antibiotics: Crystal Structure Of D-Ala-D-Ala Peptidase inhibited by a novel bicyclic phosphate inhibitor | | Descriptor: | 2-[(DIOXIDOPHOSPHINO)OXY]BENZOATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-13 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward better antibiotics: crystallographic studies of a novel class of DD-peptidase/beta-lactamase inhibitors
Biochemistry, 43, 2004
|
|
1PWC
 
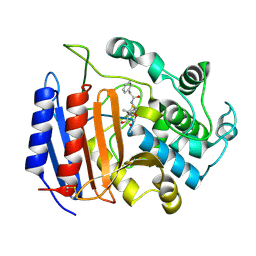 | | penicilloyl acyl enzyme complex of the Streptomyces R61 DD-peptidase with penicillin G | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, OPEN FORM - PENICILLIN G | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1SCW
 
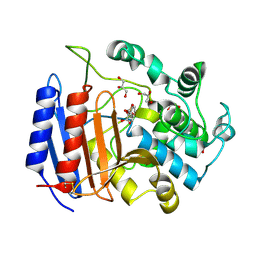 | | TOWARD BETTER ANTIBIOTICS: CRYSTAL STRUCTURE OF R61 DD-PEPTIDASE INHIBITED BY A NOVEL MONOCYCLIC PHOSPHATE INHIBITOR | | Descriptor: | (2Z)-3-{[OXIDO(OXO)PHOSPHINO]OXY}-2-PHENYLACRYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Silvaggi, N.R, Kaur, K, Adediran, S.A, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Toward Better Antibiotics: Crystallographic Studies of a Novel Class of DD-Peptidase/beta-Lactamase Inhibitors.
Biochemistry, 43, 2004
|
|
1PWD
 
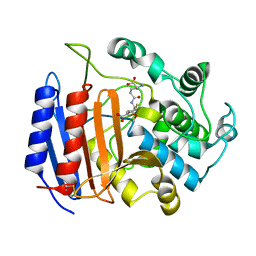 | | Covalent acyl enzyme complex of the Streptomyces R61 DD-peptidase with cephalosporin C | | Descriptor: | (2R)-5-(acetyloxymethyl)-2-[(1R)-1-[[(5R)-5-azanyl-6-oxidanyl-6-oxidanylidene-hexanoyl]amino]-2-oxidanylidene-ethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase precursor | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PW1
 
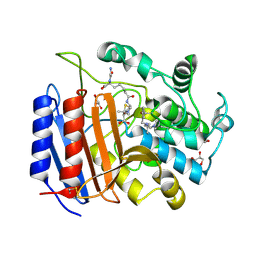 | | Non-Covalent Complex Of Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2S,5R,6R)-6-{[(6R)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, FORMYL GROUP, ... | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PWG
 
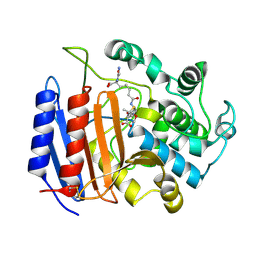 | | Covalent Penicilloyl Acyl Enzyme Complex Of The Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-carboxy-6-(glycylamino)hexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.074 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
2ILP
 
 | |
2IMB
 
 | |
2FUC
 
 | | Human alpha-Phosphomannomutase 1 with Mg2+ cofactor bound | | Descriptor: | MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUE
 
 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2IMA
 
 | |
2IMC
 
 | |
2OJR
 
 | |
3BOK
 
 | |
3BON
 
 | |
3BOO
 
 | |
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
5IHS
 
 | | Structure of CHU_2103 from Cytophaga hutchinsonii | | Descriptor: | Endoglucanase, glycoside hydrolase family 5 protein | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Periplasmic Cytophaga hutchinsonii Endoglucanases Are Required for Use of Crystalline Cellulose as the Sole Source of Carbon and Energy.
Appl.Environ.Microbiol., 82, 2016
|
|
4LZL
 
 | |
8TN3
 
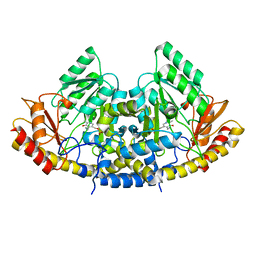 | |
8TN2
 
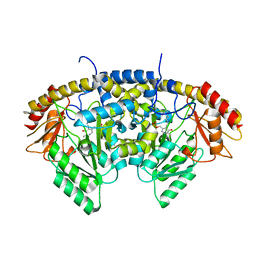 | |
5DJ3
 
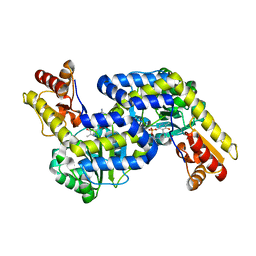 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP with D-Arginine Bound | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-D-arginine, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
5DJ1
 
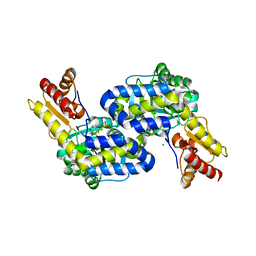 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP Holoenzyme | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
