2IFB
 
 | |
1IFB
 
 | |
3OKR
 
 | |
4XT7
 
 | |
4XT6
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with the tetrahydropteridine ring of tetrahydrofolate (THF) | | Descriptor: | (6S)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Rv2671 | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT5
 
 | |
4XT4
 
 | | Crystal structure of Rv2671 from Mycobacteirum tuberculosis in complex with dihydropteridine ring of dihydropteroic acid | | Descriptor: | 2-amino-6-methyl-7,8-dihydropteridin-4(3H)-one, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT8
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with NADP+ and trimetrexate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
5KVU
 
 | |
3F4B
 
 | |
2HH7
 
 | |
1M1M
 
 | |
8GKF
 
 | |
3SAD
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(2-mehtylphenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(2-methylphenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-02 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
6MJ1
 
 | |
6PK2
 
 | | CRYSTAL STRUCTURE OF THE CARBOXYLTRANSFERASE SUBUNIT OF ACC (ACCD6) IN COMPLEX WITH INHIBITOR QUIZALOFOP-P derivative FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | 2-{4-[(6-fluoro-1,3-benzothiazol-2-yl)oxy]-2-hydroxyphenyl}-N-methylacetamide, Propionyl-CoA carboxylase subunit beta | | Authors: | Reddy, M.C.M, Nian, Z, Michele, T.C.B, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Elucidating the Inhibition and specificity of binding of
herbicidal aryloxyphenoxypropionates derivatives to Mycobacterium tuberculosis carboxyltransferase
domain of acetyl-coenzyme A(AccD6).
To Be Published
|
|
6P7U
 
 | |
1QPQ
 
 | | Structure of Quinolinic Acid Phosphoribosyltransferase from Mycobacterium Tuberculosis: A Potential TB Drug Target | | Descriptor: | QUINOLINATE PHOSPHORIBOSYLTRANSFERASE, QUINOLINIC ACID, SULFATE ION | | Authors: | Sharma, V, Grubmeyer, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Mycobacterium tuberculosis: a potential TB drug target.
Structure, 6, 1998
|
|
1QPO
 
 | |
7S8W
 
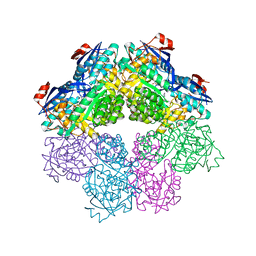 | | Amycolatopsis sp. T-1-60 N-succinylamino acid racemase/o-succinylbenzoate synthase R266Q mutant in complex with N-succinylphenylglycine | | Descriptor: | MAGNESIUM ION, N-succinyl-L-phenylglycine, N-succinylamino acid racemase/O-succinylbenzoate synthase, ... | | Authors: | Truong, D.P, Rousseau, S, Sacchettini, J.C, Glasner, M.E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Second-Shell Amino Acid R266 Helps Determine N -Succinylamino Acid Racemase Reaction Specificity in Promiscuous N -Succinylamino Acid Racemase/ o -Succinylbenzoate Synthase Enzymes.
Biochemistry, 60, 2021
|
|
7RUS
 
 | |
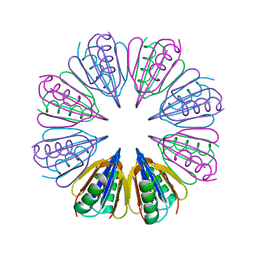
7RUR
 
 | |
6APZ
 
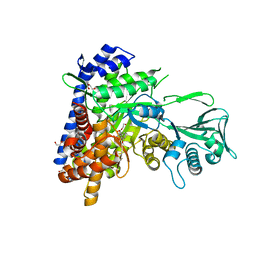 | |
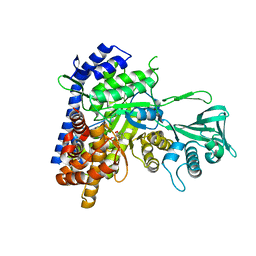
6ASU
 
 | |
6AU9
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with dioxine-phenyldiketoacid | | Descriptor: | (2Z)-4-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-hydroxy-4-oxobut-2-enoic acid, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2017-08-31 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
