9B1R
 
 | | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1 | | Authors: | Park, H, Youn, B, Park, D.J, Puthanveettil, S.V, Kang, C. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 (VSR1) from Arabidopsis thaliana.
Sci Rep, 14, 2024
|
|
4W4X
 
 | | JNK2/3 in complex with 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-fluorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4V
 
 | | JNK2/3 in complex with 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4Y
 
 | | JNK2/3 in complex with 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4W
 
 | | JNK2/3 in complex with N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide | | Descriptor: | N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
8EFJ
 
 | |
6N2U
 
 | |
1YPO
 
 | |
1YPQ
 
 | |
8VMH
 
 | |
8VO4
 
 | |
8VS0
 
 | |
8VT6
 
 | |
8VMM
 
 | |
8VNX
 
 | |
8VTF
 
 | |
8VWM
 
 | |
1N4E
 
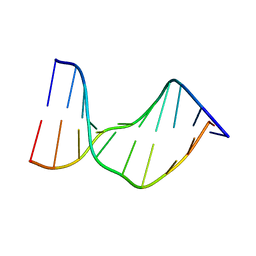 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer containing a cis-syn thymine dimer.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1RF6
 
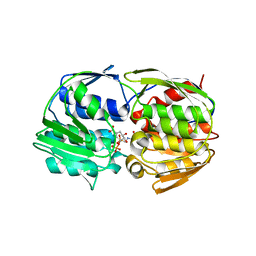 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RF4
 
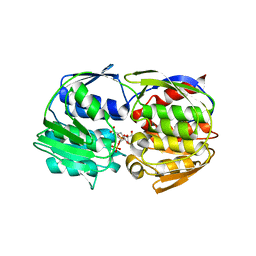 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RF5
 
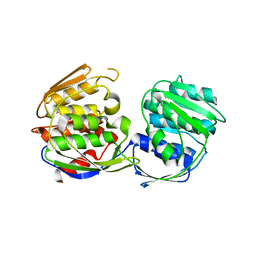 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
3O3U
 
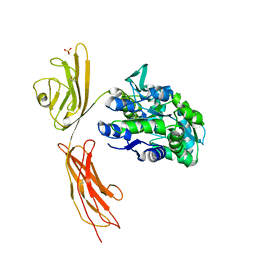 | | Crystal Structure of Human Receptor for Advanced Glycation Endproducts (RAGE) | | Descriptor: | Maltose-binding periplasmic protein, Advanced glycosylation end product-specific receptor, SULFATE ION, ... | | Authors: | Park, H, Boyington, J.C. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | The 1.5 A crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding.
J.Biol.Chem., 285, 2010
|
|
1SM5
 
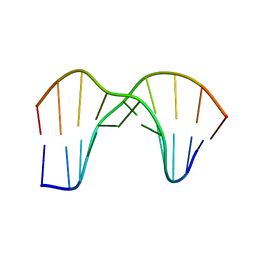 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3RF3
 
 | |
5BTM
 
 | |
