6IBL
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
7B5J
 
 | | Anti-CRISPR associated (Aca) protein, Aca2 | | Descriptor: | Anti-CRISPR associated (Aca) protein, Aca2, GLYCEROL | | Authors: | Usher, B, Birkholz, N, Fineran, P.C, Blower, T.R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the anti-CRISPR repressor Aca2.
J.Struct.Biol., 213, 2021
|
|
9GBK
 
 | | Blm10-20S proteasome complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome subunit alpha type-1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-07-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis
To Be Published, 2024
|
|
9D5D
 
 | |
2Y04
 
 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST SALBUTAMOL | | Descriptor: | BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, HEGA-10, ... | | Authors: | Warne, A, Moukhametzianov, R, Baker, J.G, Nehme, R, Edwards, P.C, Leslie, A.G.W, Schertler, G.F.X, Tate, C.G. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor
Nature, 469, 2011
|
|
2XDB
 
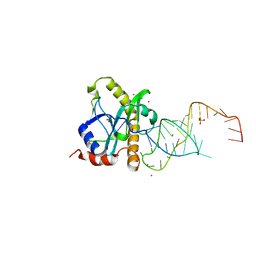 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | COBALT (II) ION, SULFATE ION, TOXI, ... | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Processed Noncoding RNA Regulates an Altruistic Bacterial Antiviral System.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XDD
 
 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | TOXI, TOXN, ZINC ION | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-30 | | Release date: | 2011-01-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Phage Abortive Infection System, Toxin, Functions as a Protein-RNA Toxin-Antitoxin Pair.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5XEW
 
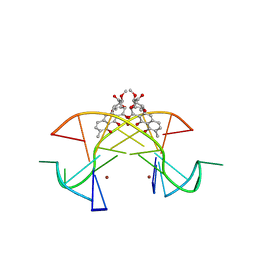 | | Crystal structure of the [Ni2+-(chromomycin A3)2]-CCG repeats complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Tseng, W.H, Wu, P.C, Hou, M.H. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel-Chromomycin Complex Resulting in Large-Scale DNA Deformation
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2KXG
 
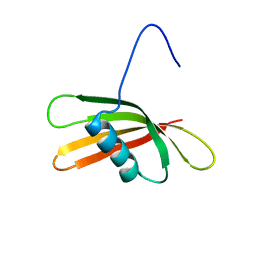 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
5XJW
 
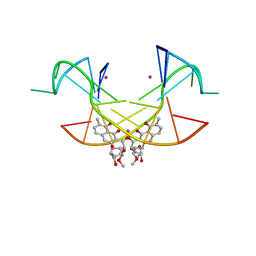 | | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Tseng, W.H, Wu, P.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal Structure of the [Co2+-(Chromomycin A3)2]-CCG repeats Complex
To be published
|
|
6ITO
 
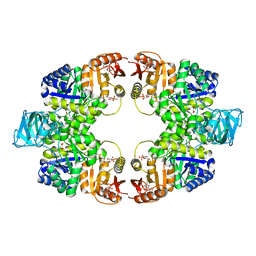 | | Crystal structure of pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, AMP and inhibitor Ribose 5-Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Mu, Y, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-11-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Pyruvate Kinase Regulates the Pentose-Phosphate Pathway in Response to Hypoxia in Mycobacterium tuberculosis.
J.Mol.Biol., 431, 2019
|
|
2CCY
 
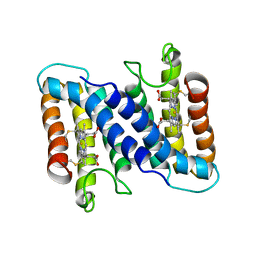 | | STRUCTURE OF FERRICYTOCHROME C(PRIME) FROM RHODOSPIRILLUM MOLISCHIANUM AT 1.67 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Finzel, B.C, Weber, P.C, Hardman, K.D, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution.
J.Mol.Biol., 186, 1985
|
|
4V25
 
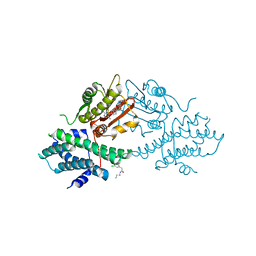 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V26
 
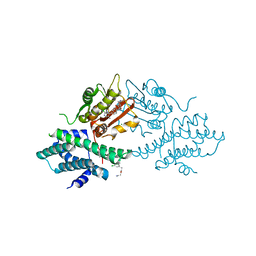 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V5M
 
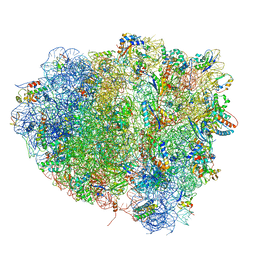 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
2XD0
 
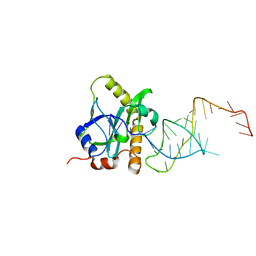 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | TOXI, TOXN | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-28 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A processed noncoding RNA regulates an altruistic bacterial antiviral system.
Nat. Struct. Mol. Biol., 18, 2011
|
|
7S0P
 
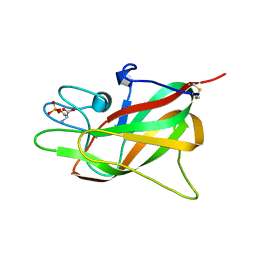 | | Crystal structure of Porcine Factor VIII C2 Domain Bound to Phosphatidylserine | | Descriptor: | Coagulation factor VIII, PHOSPHOSERINE | | Authors: | Peters, S.C, Childers, K.C, Wo, S.W, Brison, C.M, Swanson, C.D, Spiegel, P.C. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Stable binding to phosphatidylserine-containing membranes requires conserved arginine residues in tandem C domains of blood coagulation factor VIII.
Front Mol Biosci, 9, 2022
|
|
4XZU
 
 | |
1QBQ
 
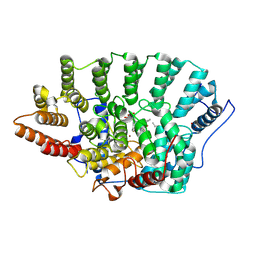 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
1QF7
 
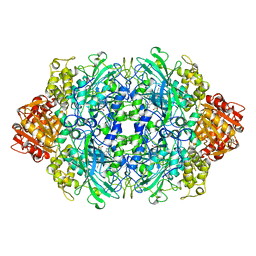 | |
4LMF
 
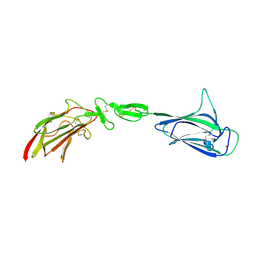 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2A8D
 
 | | Haemophilus influenzae beta-carbonic anhydrase complexed with bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A8C
 
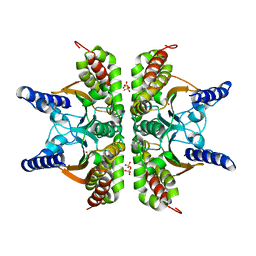 | | Haemophilus influenzae beta-carbonic anhydrase | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A9E
 
 | |
2AEB
 
 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
